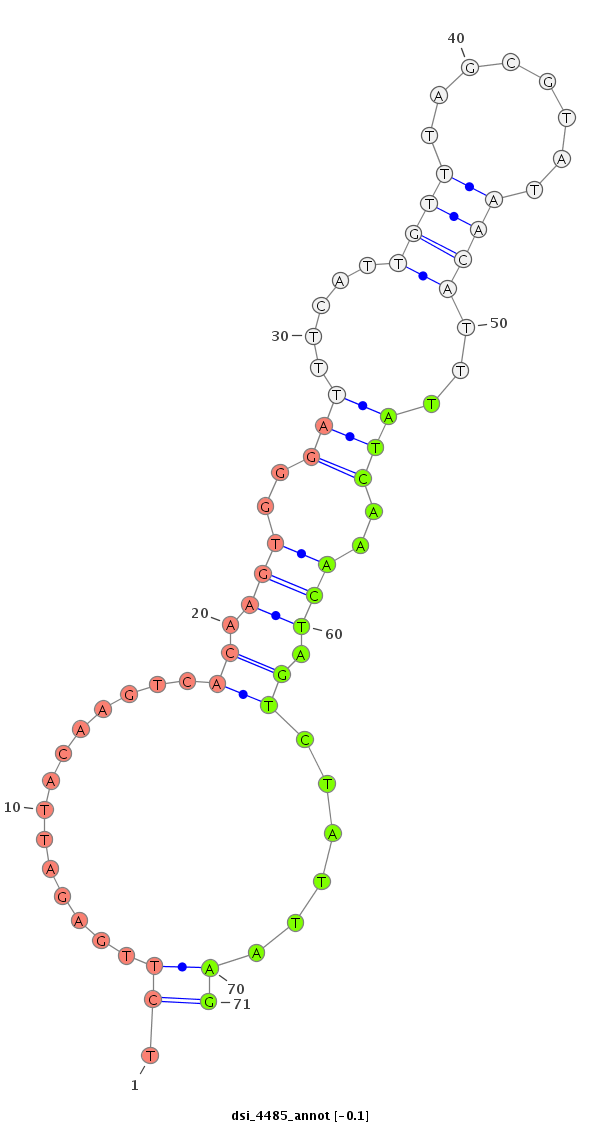
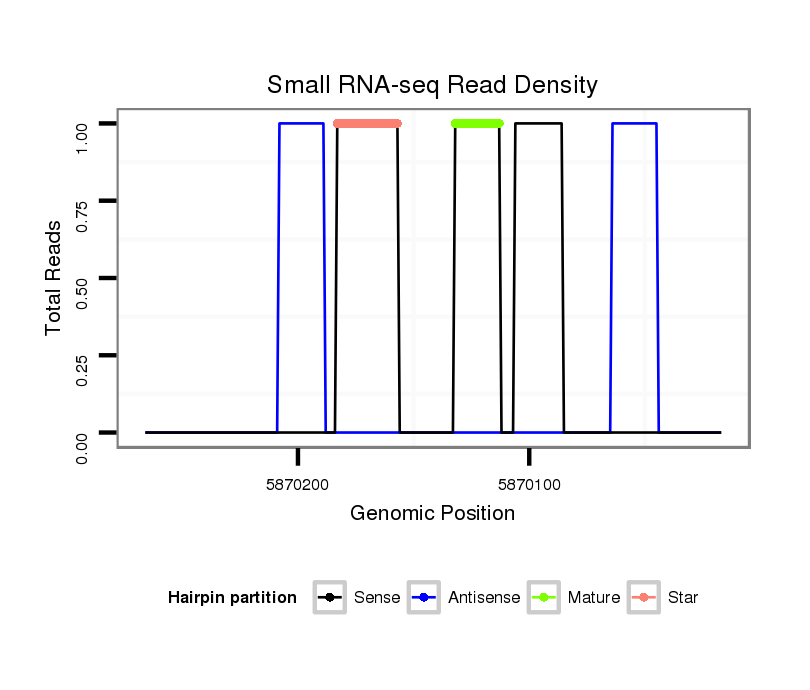

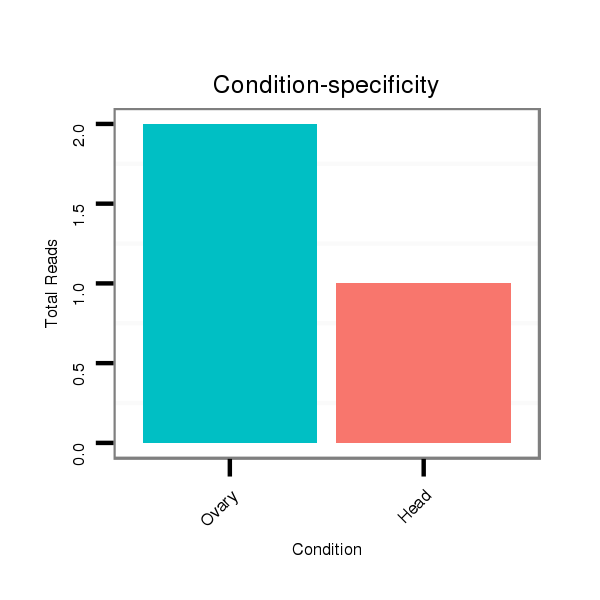
ID:dsi_4485 |
Coordinate:2r:5870067-5870216 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -7.9 | -7.0 |
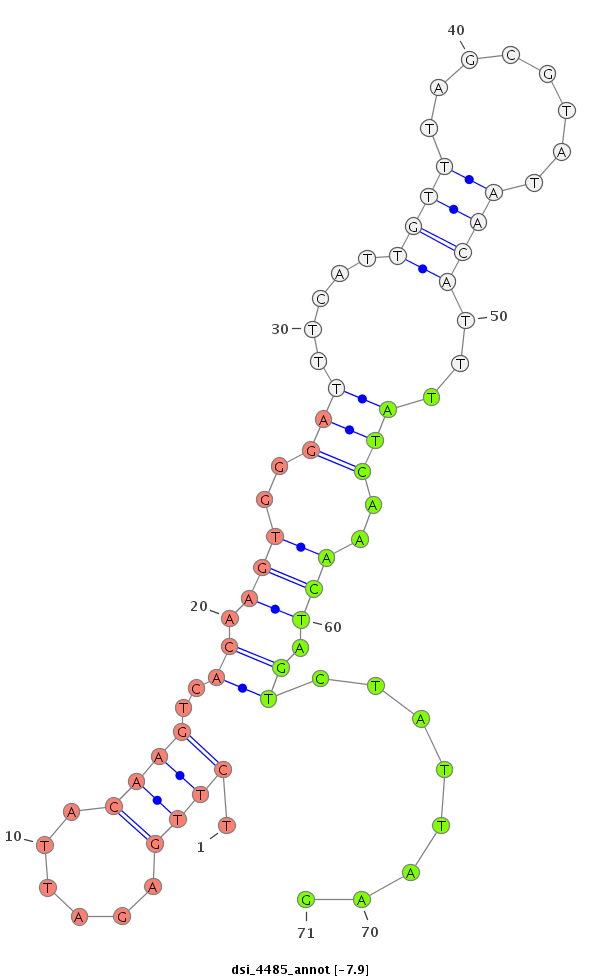 |
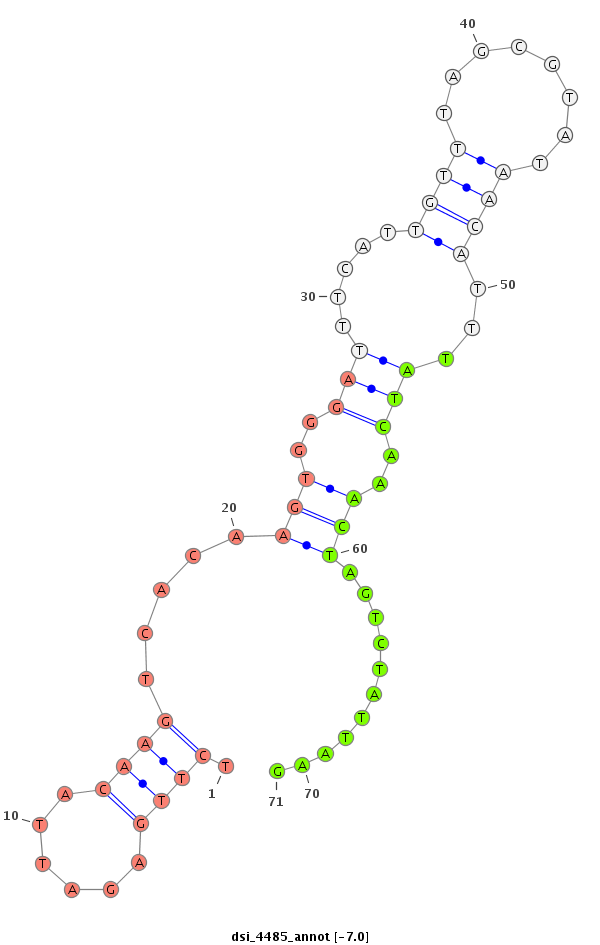 |
exon [2r_5869671_5870066_-]; CDS [2r_5869671_5870066_-]; intron [2r_5870067_5870589_-]
No Repeatable elements found
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TTACTAGCTATTATAGTTCGAAATGCAGCGATTTTTTGTATTTGTATCTTGAAAAACAGAGCCCAAAAGTATGTATCTCGTTATCTTGAGATTACAAGTCACAAGTGGGATTTCATTGTTTAGCGTATAACATTTATCAAACTAGTCTATTAAGCATGGATCGAACATACAAGTAACTCTCATCCAATCCTTGCTTGCAGGCCATCATGACATCCACCAGCGATAAGCGTGAGCATCTGTACAAGATTCT ***********************************************************************************.((..............((.(((..(((.....((((........))))...)))..))).))......))************************************************************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
M023 head |
SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................TATCAAACTAGTCTATTAAG................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .........................................................................................GATTACAAATCAGACGTGGGA............................................................................................................................................ | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ...................................................................................TCTTGAGATTACAAGTCACAAGTGGGA............................................................................................................................................ | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................................TCGAACATACAAGTAACTCTC..................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................TCCTTGCTTGCAGGCC............................................... | 16 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
|
AATGATCGATAATATCAAGCTTTACGTCGCTAAAAAACATAAACATAGAACTTTTTGTCTCGGGTTTTCATACATAGAGCAATAGAACTCTAATGTTCAGTGTTCACCCTAAAGTAACAAATCGCATATTGTAAATAGTTTGATCAGATAATTCGTACCTAGCTTGTATGTTCATTGAGAGTAGGTTAGGAACGAACGTCCGGTAGTACTGTAGGTGGTCGCTATTCGCACTCGTAGACATGTTCTAAGA
************************************************************************************************.((..............((.(((..(((.....((((........))))...)))..))).))......))*********************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902009 testis |
M023 head |
SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................................TTGTCCGGTAGTACTGTAGG................................... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................CTCGGGTTTTCATACATAGA............................................................................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................................................................................GTAGTACTGTAGGTGGTCGC............................ | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................AACTCTAATTTTCAGAGTTC................................................................................................................................................. | 20 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................TCGCACCTAGCTTGT................................................................................... | 15 | 1 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 |
| .................................................................................................CAGTCTTCACCCTAAA......................................................................................................................................... | 16 | 1 | 19 | 0.16 | 3 | 0 | 0 | 3 | 0 |
| ...........................................................................................................................................................TCCTTAGCTTGTATGTT.............................................................................. | 17 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 01:40 AM