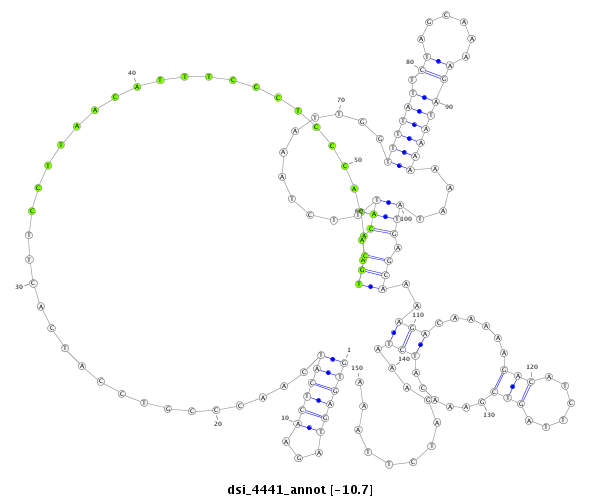
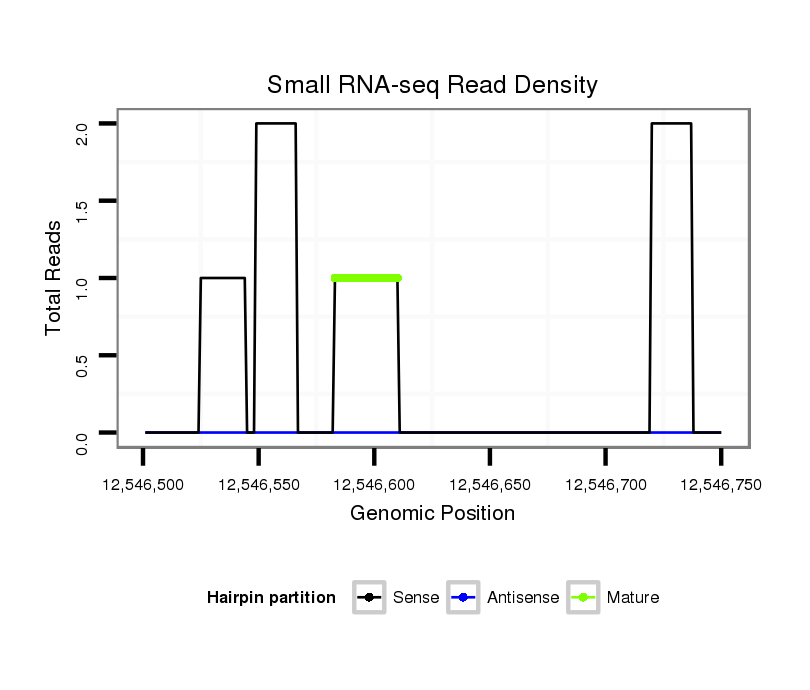
| droSim2 |
3l:12546501-12546750 + |
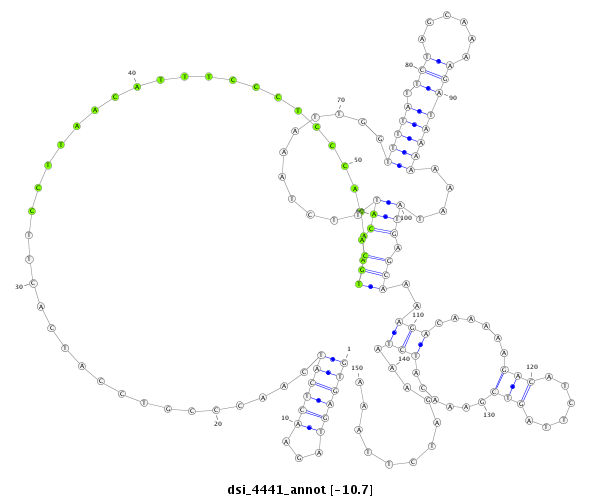
dsi_4441 |
AGCCGCCG------------------------CATCT------------------------------GGAGCCAGCTTTATGGGCTTCAGTTTGCCCCCTGGAAGTGAGTAGAAC-------------TCATCAACC-----------------------------------------CGTCCATCACT----TCCTTAACATTTCCCTCCCA---CAATGCACATT------------------TCTAAATTGGTTTT----ATTCTAGCAAAAGATAA--------A---------------------AAAATATGAGCAAAAG-ACAAA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGACATCTT------AGTC-----------------------------------------------GA-----------------------------AA----------------------------------CATCT------AAAGATCTTAAATGTTATCAAGAATTTAATTGA----------------AGTATCATGAAGGA------------------------C----------C--TTTTGTTTTCGGA |
| droSec2 |
scaffold_0:5028851-5029114 + |
|
AGCCGCCG------------------------CATCT------------------------------GGTGCCAGCTTTATGGGCTTCAGCTTGCCCCCTGGAAGTGAGTAGAAC-------------TCATCAACC-----------------------------------------CGTCTATCACT----TCTTTAACATTTCCCTCCCA---CAATGCACATT------------------TCTAAATTGGTTTT----ATTCTAGCAAAAGATAA--------A---------------------AAAATATAAGCAAAAGGACAAGAAATTT-ATG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AACATCTTTAGAATAGTC-----------------------------------------------GA-----------------------------AA----------------------------------CATCT------AAAGATCTTAAATGTTATCAAGAATTTAATTGA----------------AGTATCATGAAGCA------------------------C----------C--TTTTGTTTTCGGA |
| dm3 |
chr3L:12844773-12845036 + |
|
AGCCGCCG------------------------CATCT------------------------------GGAGCCAGCTTTATGGGCTTCAGCTTGCCCCCTGGAAGTGAGTAGAAC-------------TCATCATCC-----------------------------------------CATTCATCACT----TTTTTAACATTTCCCTCCCA---CAATGCACATT------------------TCTAAATTGGTTTT----ATTCTAGCAAAAGATAA--------A---------------------AAAATATGAGCAAAAGGACAAGAAAATT-ATG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AACATCTTTAAAATAGTC-----------------------------------------------GA-----------------------------AA----------------------------------CATCT------AAAGATCTAAAATGTTATCAAGAATTTAATTGA----------------AGTATCATGAACCA------------------------C----------C--TTTTGCTTTCGGA |
| droEre2 |
scaffold_4784:12857236-12857517 + |
|
AGCCGCCG------------------------CATCT------------------------------GGAGCCAGCTTTATGGGCTTCAGCCTGCCCCCTGGAAGTGAGTAGATA-------------TCGTCAACC---------------------CAGTAGTTG--------AACTCCCCCTC--------ACTTAACATTCCCTTCCCA---CAATGCACTTT------------------TCTAAATTGGTTTT----ATTTTAGCAGAAGAGAA--------A---------------------ATAACATAAGCAAAGCGACAAGAAAATT-ATA-TCTTAAAAGAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAACATCTTTTAGCAAGTC-----------------------------------------------GA-----------------------------AA----------------------------------CATCT------AAAGATCTAATACGTTATCAAGAATTTCA---A----------------ATGAATATGAAAAA------------------------C----------C--TTCTGCTTTAGGA |
| droYak3 |
3L:12920454-12920730 + |
|
AGCCGCCG------------------------CATCT------------------------------GGAGCCAGCTTTATGGGCTTCAGCTTGCCCCCTGGAAGTGAGTAGAGC-------------TCATCAACC---------------------TAGTAGTTG--------AACTCCCCATT--------AATTAACATTCCCTTTCCA---CAATGCACTTT------------------TCTAAATTGGTTTT----ATTTTAGCAGAAGAAAA--------A---------------------ATAACATAAGCAAAGGGAGAAGAAAATT-ATG-AATTAGAAGAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAACCATCTTTAAG-----C-----------------------------------------------AA-----------------------------AA----------------------------------CAATT------TAAGATCTAAAATTTTATCAAGAATTTAA---A----------------AATAATATAAAATA------------------------C----------C--TTCTGCCTTAGAA |
| droEug1 |
scf7180000409466:4943253-4943558 + |
|
AGCTGCTG------------------------CATCT------------------------------GGAGCTAACTTTATGGGTTTCAGCTTGCCACCAGGAAGTGAGTAGAAC-------------TCATCCTTA---------------------AACTAATTA--------TATTTCCCATCATT----TACTTAACATTCCCCTCCTA---TTATGCACATT------------------ATTAAATTGGTTTT----ATTATTGCAGAAGAAAA--------A---------------------AC---AGAATCATAAAG-----TAA------------------------ATTG-AAGAACTAATTCAA----G----------------------------------------------------------------------------------------------------------------------------------------------------------AA-----------------------CTATTTGA----AATA----------TATTTAAGG-------AAGATTTAA--------------------AATGGAAGCTTTATATTTTAT-------------------------------------TTGGCATT----------------------------------------AAG-ATGCAATATATAATCAAGAAGCAT-TAAGAAGTTTCAAGCATGATTATAGA |
| droBia1 |
scf7180000302529:773423-773713 - |
|
AGCAGCTG------------------------CATCT------------------------------GGAGCCAGCTTTATGGGCTTCAGCTTGCCCCCAGGAAGTGAGTAAAAC-------------TCATCCTCC---------------------ACGTAGTTC--------TTAC-TCCTTCATT----GAATTAACATTCCCTTCCCA---TAATGCACTTT------------------GTAAAATTGGTTT-----ATTCTTGCAGAAGATAA--------A---------------------GAAACGAAAGAATAAGAAGAAAGGGATGCAAGAAATTAGATGCATGCTAA------------AC----TTAAAGTAATGAA---------------------------------CACGAATTTACCAAACCAGTTTAA-----------------AAAATCATGAAATCTCG---------------------------------------------------------------CAAACTAACCA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAAA------------------------C----------C--TTTAATATTATTT |
| droTak1 |
scf7180000415251:204776-205070 - |
|
AGCCGCTG------------------------CATCT------------------------------GGAGCCAGCTTTATGGGCTTCAGCCTGCCACCAGGAAGTGAGTATAAC-------------TCATCCATC---------------------AAGTAGTTA--------TTATCCTCATCATT----TATCTCACATTCCTCTCCCA---TATTGCACTTT------------------ATTAAATTGGTTTA----ATTCCCGCAGAAGAAAA--------A---------------------ATAACAAAAGCATAAGGAGAGGAAAATGCAAGAATTC-------------------------CT----TTAAACCTATCTA---------------------------------TATAAATTCATTAAAGCCGTTGAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AACAATCAAGAAGCTTTAAATAGTAT-------------------------------------TTCCCATA----------------------------------------AAG-ACGTAAAAACTAA-------------------------TACATTTTTAGCA |
| droEle1 |
scf7180000491255:1240137-1240459 + |
|
AACCGCTG------------------------CAGCC------------------------------GGAGCCAGCTTCATGGGTTTCAGCTTGCCGCCAGGAAGTGAGTTTAAT-------------TTATTTACC---------------------ATGATGTTG--------CATCGACCATAATT----AATTTAACATTTCCCTCCTT---TAATGCACCTT------------------ACTAAATTGATTAA----ACTTTTGCAGAAGATAA--------A---------------------ATAACAAAAGCATAAGGACATGAAAATCCAAAAAA-TAGCAGCATGAAATTAATTA-----------------------------------------------------GCTT------------------------------------------------------------------------------------------------------------------------------------------------TAA----AATGTTTTACGATTTTTGCAATG-------AAAATCTGAAAGTTAAAAAAGGGATTCAAGCAAGAAGC---------------ATCAAATATTAACA---------------------ACTAATC------------------------------------AAGAAGAA------------------------T----------C--GATTAATTACTAA |
| droRho1 |
scf7180000766682:21928-22254 + |
|
AGCCGCTG------------------------CATCC------------------------------GGAGCCAGCTTTATGGGTTTCAGCTTGCCGCCAGGAAGTGAGTTTAAA-------------GTATTTATC---------------------ATGTAGTTG--------TAAATCAATTCATT----TATTTAACATTCCCCTCCTA---TAATGCACTTT------------------ATTAAATTGATTCA----ACTTTTACAGAAGAAAA--------A---------------------ATAACAAATGCTTAAGGAGATGAAAATCCATGAAATTAACAGCA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGAACATCTCTAAATTAATCA--AAATCTTCTTGA----TATG----------TTTACGATG-------AAGATCTGAAAACTAATGAAAGGTTTTAAGAAAG-TT-------------AGCATG----------------------------AAAATTTCATC------------------------------------AAGAGGCA------------------------TTAAAG-----A--AATAGCATAACGA |
| droFic1 |
scf7180000453839:162629-162899 - |
|
AGCCGCTG------------------------CATCC------------------------------GGAGCCAGCTTCATGGGCTTCAGCTTGCCGCCAGGAAGTAAGTATAAT-------------CCATACACA---------------------TAGTAGTTT--------TTACTCCGATCATG----TCTTTCACATTCCCTTTCCT---AAACGCACTTT------------------ACTAAATTGGTTTA----C--CTTACAGAAGAATA--------ATCGAAGCATACGGCTAACAAAGTAGAAAATAAT-AAGGGCAGGAAAATT-TTG-------------------------------------------------------------------------------------------------------------------------AAATAATCT--------------------------------------------------------------------------A---ATGTG-----------------------------------------------GAAAGATCTGAAAACTAA---------TCAAACAAAAAT-----------------------------------------AGCTCTACAAACAAATC-------------------------------------------------------------------------------------------AAGA |
| droKik1 |
scf7180000302272:491682-491981 + |
|
AACTGCCG------------------------CTTCG------------------------------GGAGTCAACTTTATGGGCTTCAGCCTGCCGCCAGGAAGTGAGTAAAGC-------------TTAAACCTA---------------------AATTAATCT--------T--CATTTATCACTTAGTTCATTATCATTTCCTTCCTTTAACTATGCACAAT------------------TTGAATTAGCTTTT----ACTTTTGCTGAAGGAGGTGGTGATAA---------------------A------AAG---AATGACAAGTAA------------------------TCAATAGGAAATGATTCAT----G----------------------------------------------------------------------------------------------------------------------------------------------------------AA-----------------------TGTATAGA----AATA----------TTT--ATTAAAAGGG------------------------AATCGAAATGGAAG-----------------------------------------------------TCATCAAGTATT----AGT----------------AGGATCATTAAAC-CCATAAAGTATAT-------------------------TAATATTATGGAA |
| droAna3 |
scaffold_13337:19742954-19743164 - |
|
TGCAGGTG---------------CAACTGGGGCCACG------------------------------GGTGTTAGCTTCTTGGGATTTAGCTTGCCGCCAGCAAGTAAGTAAATT-------------CGAAT--CT-----------------------------------------TGTTTTTCATT------TTTAAAATTCATTTTCTT---TGTCGCACATT------------------ATTCATTTTAA------------------------------------------------------------------------------------------------------------------------------AACCATTTCACTGCAAATAGCACCAAGAAAACGGTCCACTATCTC---------------------AAACATGTAGTCTCGAGAATTTCAAGAAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAATAGCCA |
| droBip1 |
scf7180000396360:931355-931619 - |
|
AACTGCGG------------------------CCTCG------------------------------GGAGTTAGCTTCTTGGGATTCAGCTTGCCGCCAGGAAGTGAGTAACTT---------------AA-AATT-----------------------------------------TATTTATTTTT----ATTTTAAAAATCATTTTATT---CGTTGCACATT------------------ATTTATTT-----------------------------------------------------------------------------------------------------------------------------TTAAACCCATCTCACTGCCAGTATCATCAAGAAAGCAGTCTAATGACTC---------------------AAACACTC-----------------------------------------------------------------------------------CATCTT------AGTTGACAAAATTTCAAGATACAATA-------------------------GCAAGATCAAACA-----------------------------------------------------------------------------ACTAATCAAGATAATAA---C----------------AATAA-CTGGGATA------------------------T----------T--TTTTGTTGCCAAA |
| dp5 |
XR_group6:11621353-11621574 - |
|
-GCCGGCG------------------------CTGCG------------------------------GGTGCCAGTTTTATGGGCTTCAGTCTGCCGCCAGGAAGTGAGTAATGCAATCCTTCAGA------------TATCCCCACACACATGTTCA---------TTACTTAT---------------------T--------------------CACGCATAAC------------------CATGATTTGGCCTTCAACTTTTTAGCGGAAAGTA----------------------------------AAAAATGCTAAAAGGCAAGCAAGTT--------------------------------------------------------------------------------------------------------------------------CCAAGAAAATCAAGAAATCAAGAGCCCCCCAC------------TCATTTTAT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTGTTATC-------------------------------------------------------------------------------------------AAAA |
| droPer2 |
scaffold_20:1546254-1546475 - |
|
-GCCGGCG------------------------CTGCG------------------------------GGTGCCAGTTTTATGGGCTTCAGTCTGCCGCCAGGAAGTGAGTAATGCAATCCTTCAGA------------CAATCCCACACACATGTTCA---------TTACTTAT---------------------T--------------------CACGCATAAC------------------CATGATTTGGCCTTC---AACTTTTTAGAGGAAA-------------------------------GTAAAAAATGCTAAAAGGCAAGCAAGTT--------------------------------------------------------------------------------------------------------------------------CCAAGAAAATCAAGAAATCAAGAGCCCCCCAC------------TCATTTTAT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTGTTATC-------------------------------------------------------------------------------------------AAAA |
| droWil2 |
scf2_1100000004511:5453279-5453625 - |
|
TGCCGCCG------------------------C------------------------------------TGCCAGTTTTATGGGCTTCAGCTTGCCACCCGGAAGTGAGTATTGAAGTCATCTCCTTAACACAGACGAATATTCCATTCATT--------------------------CAATCATTATT----CATTTAATTTTTATTT----------TGCTTAGCCAAAACGGAAAGCACTTGACTC---AGATCTT----GATCTAACAAAAGGTTC--------A----AACATTGGACTCAC---------------------------AGTT--------------------------------------------------------------------------------------------------------------------------CC------------------------CCCTCCTCCATGTATTAGTCGTTTTATTTGTTTTGGCCCAAAAAAAAAAATAATGTTTT------AGCA-----------------------------------------------GG-----------------------------AA----------------------------------ATTCAAGCCAGCACGATATTAT-GGTTATAAATAAATTAATTTAAATTCTTGATATTATATATATTATAAAA--------------------------G----------T--TGTTGTTCATTGA |
| droVir3 |
scaffold_13049:22677310-22677495 - |
|
AGGCGGTG------------------------CGGCT------------------------------GCAGCAAACTTTATGGGCTTCAGCTTGCCACCTGGAAGTGAGTATTTG-------------ATACAGACA---------------------CATCA--------TTATT--AACCCATCATT---------------CATA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TT-TTTGT-------------------------------------TGTTCAGATTAAATGATCAAGTC-------------------------ACGTTTAAAACAAATT---------AGGCAATGCAAGCGC------------------------------------------------------A-------TCAAATTGG----------------AGTTTTCTCAATGA------------------------G----------C-----------CGTA |
| droMoj3 |
scaffold_6680:17296510-17296587 - |
|
AATGTCTGCTGCCAACGCAGGAGGAAGTA---GTGCT------------------------------GCGGCTAATTTCATGGGCTTCAGCTTGCCACCTGGAAGTTAGTA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droGri2 |
scaffold_15110:20855078-20855336 + |
|
AGCTGCTG------------------------GTGCTGGTGCCAGCGGTGCCGCTGGCAGTGCGGCCTCCGTCAACTTTATGGGCTTCAGCTTGCCACCAGGAAGTGAGTATTTG-------------ACATATAT-----------------GCCTACA--AAGCCTTATTTAT-TTAATCCATCATT---------------CATT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTATTTGT-------------------------------------TGTCCAGACTAATTGATCAAAACA----------TTA--ATGG---------------AAAATT---------AAGCAAAATAACCGT---------------ATGC--------AATCT---CAGCAAGATCTTATCCGTTACGAATAA----ATTGG----------------TGTGTTATAA-------------------------------------------TTTTTTTTCTTG |