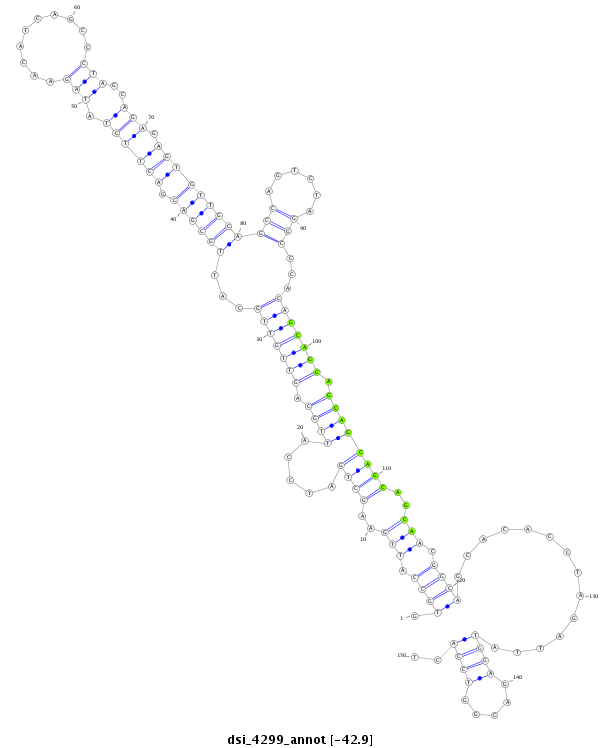
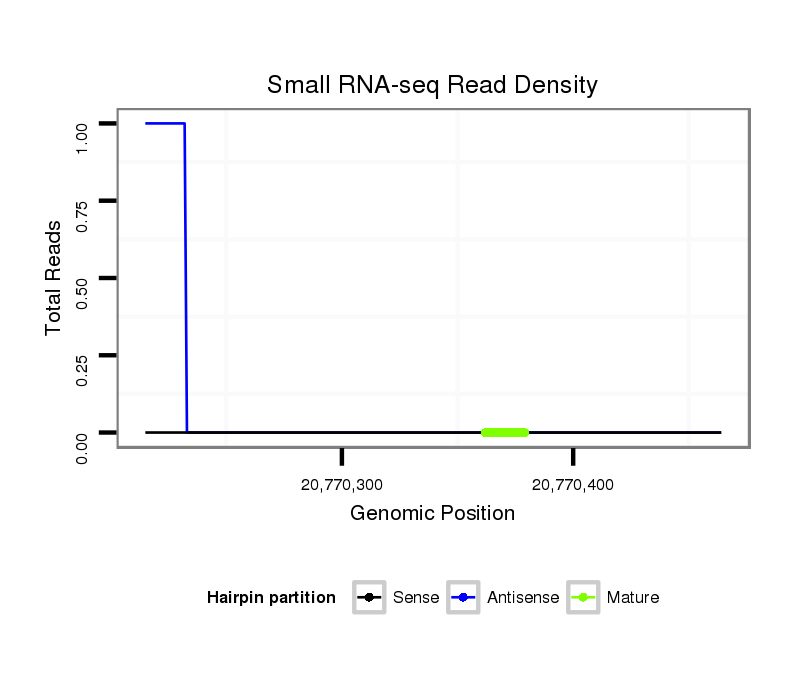
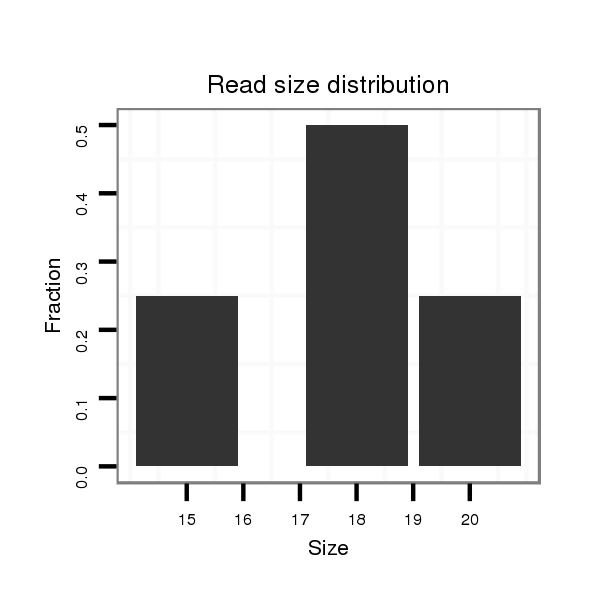
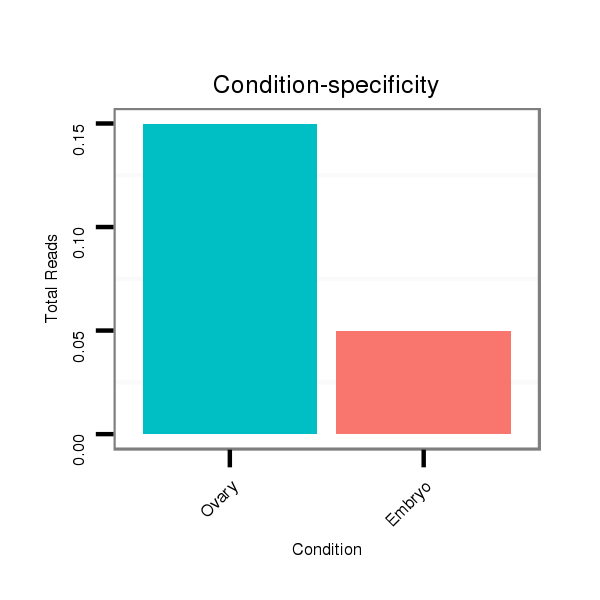
| ACAGCAAC---------A------------GC---AGCAACAGCAGCAACAGCA------------------GC---AGCAGCAAC-AGCA---------GCAG---CA----------------------ACAGCAGCAG-CAG-------------------CAA--CAGCAG------------AAGCAGCAAC----------------------------------------------AGCA-------------------------------------------------------------------GCAACAGCAACAGCAGCAACAGCAGCAGCAAC-AACAGCAGCAGCAACAGCAACAGCAG------------------CAGCAGCAGCAGCAACAGCAGCAGCAGC--------------------------A--------ACAGCAG-CAGCAGCAGCAATAG--CGGCAACAGCCAC-----------------------------------------A---------------------------------------------------------GCA------TCAGCA-ACAGCATCAGCAACAG---CAG---------------CAGC | Size | Hit Count | Total Norm | Total | M046
Female-body | V041
Embryo | V049
Head | V056
Head | M060
Embryo | V110
Male-body |
| ........................................................................................................................................AGCAG-CAG-------------------CAA--CAGCAG------------AAGCAGCA..................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................................................AA--CAGCAG------------AAGCAGCAAC----------------------------------------------AGCA-------------------------------------------------------------------.............................................................................................................................................................................................................................................................................................................................................. | 22 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................AACAGCAGCAGCAACAGCAA......................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................A--CAGCAG------------AAGCAGCAAC----------------------------------------------AGC.................................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................GCAGCAGC--------------------------A--------ACAGCAG-CAGCAGC................................................................................................................................................................................... | 23 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................CAACAGCAACAGCAG------------------CAGCAGC.......................................................................................................................................................................................................................................................... | 22 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................GCAGCAACAGCAACAGCAG------------------CA............................................................................................................................................................................................................................................................... | 21 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................CAACAGCAACAGCAGCAACA......................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................AACAGCAACAGCAG------------------CAGCAGCA......................................................................................................................................................................................................................................................... | 22 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................................................AGCAGCAGCAACAGCAACA....................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................ACAGCAGCAGCAAC-AACAGC....................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................AGCAGCAAC-AACAGCAGCAGC................................................................................................................................................................................................................................................................................................. | 21 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................GCAGCAACAGCAACAGCAG------------------CAGC............................................................................................................................................................................................................................................................. | 23 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................CAG-------------------CAA--CAGCAG------------AAGCAGCA..................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................CAACAGCAACAGCAGCAACAG........................................................................................................................................................................................................................................................................................................................ | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................AGCAACAGCAGCAGCAAC-AACAGCAGC.................................................................................................................................................................................................................................................................................................... | 27 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................................GCA------------------GC---AGCAGCAAC-AGCA---------GC.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................AGCAGCAACAGCAACAGCA.................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................AGCA------------------GC---AGCAGCAAC-AGCA---------GCA................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................................GCA------------------GC---AGCAGCAAC-AGCA---------GCA................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 21 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................CAGCAGCAACAGCAGCAGCAAC-............................................................................................................................................................................................................................................................................................................. | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................................AACAGCAGCAACAGCA------------------GC---AGC........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...............................C---AGCAACAGCAGCAACAGCA------------------GC---AGCAG...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................GCA------------------GC---AGCAGCAAC-AGCA---------GCAG---CA----------------------AC................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................................................................................................................................................ACAGCAACAGCAGCAACAGCAGC.................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................................AG------------------CAGCAGCAGCAGCAACAGCAGCAGCA....................................................................................................................................................................................................................................... | 28 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....CAAC---------A------------GC---AGCAACAGCAGCAACAGC................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................CAGCAACAGCAG------------------CAGCAGCAGCAGCAAC................................................................................................................................................................................................................................................. | 28 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................GCAACAGCAACAGCAGCAACAGCAG..................................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................GCAG---CA----------------------ACAGCAGCAG-CAG-------------------.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................AGCAGCAAC-AACAGCAGCAGCAACAGC........................................................................................................................................................................................................................................................................................... | 27 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................AGCAAC-AACAGCAGCAGCAACAGC........................................................................................................................................................................................................................................................................................... | 24 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................................AGCAGCAGCAAC-AACAGCAGCAGCAA............................................................................................................................................................................................................................................................................................... | 26 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................................................AAC----------------------------------------------AGCA-------------------------------------------------------------------GCAACAGCAACAGCA............................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................AGCAGC--------------------------A--------ACAGCAG-CAGCAGCA.................................................................................................................................................................................. | 22 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................................AG------------------CAGCAGCAGCAGCAACAGC.............................................................................................................................................................................................................................................. | 21 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................ACAGCAGCAGCAAC-AACA......................................................................................................................................................................................................................................................................................................... | 18 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................A-------------------------------------------------------------------GCAACAGCAACAGCAGCAACAGCAG..................................................................................................................................................................................................................................................................................................................... | 26 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................CAACAGCAACAGCAG------------------CAGCAGCAGCAGC.................................................................................................................................................................................................................................................... | 28 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................C-AACAGCAGCAGCAACAGCA.......................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................GCAGCAAC----------------------------------------------AGCA-------------------------------------------------------------------GCAACAGCAACAGCA............................................................................................................................................................................................................................................................................................................................... | 27 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................AACAGCAGCAACAGCAGCAGCAAC-............................................................................................................................................................................................................................................................................................................. | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CA-ACAGCATCAGCAACAG---C..................... | 19 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................AGCAAC-AGCA---------GCAG---CA----------------------ACA.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 19 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................CAACAGCA------------------GC---AGCAGCAAC-AGCA---------GCA................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 26 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................ACAGCAGCAACAGCAGCAGCAAC-............................................................................................................................................................................................................................................................................................................. | 23 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................AC-AACAGCAGCAGCAACAGCAACA....................................................................................................................................................................................................................................................................................... | 24 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................CA---------GCAG---CA----------------------ACAGCAGCAG-CAG-------------------C................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................CAG------------------CAGCAGCAGCAGCAACAGCAGC........................................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CATCAGCAACAG---CAG---------------CAGC | 19 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |