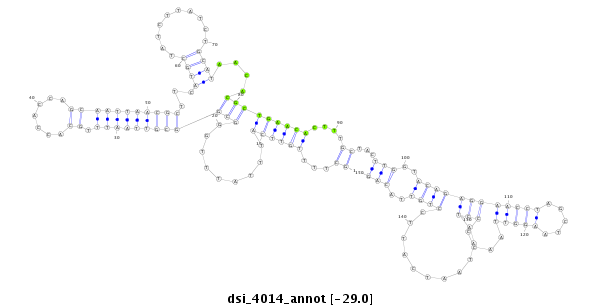
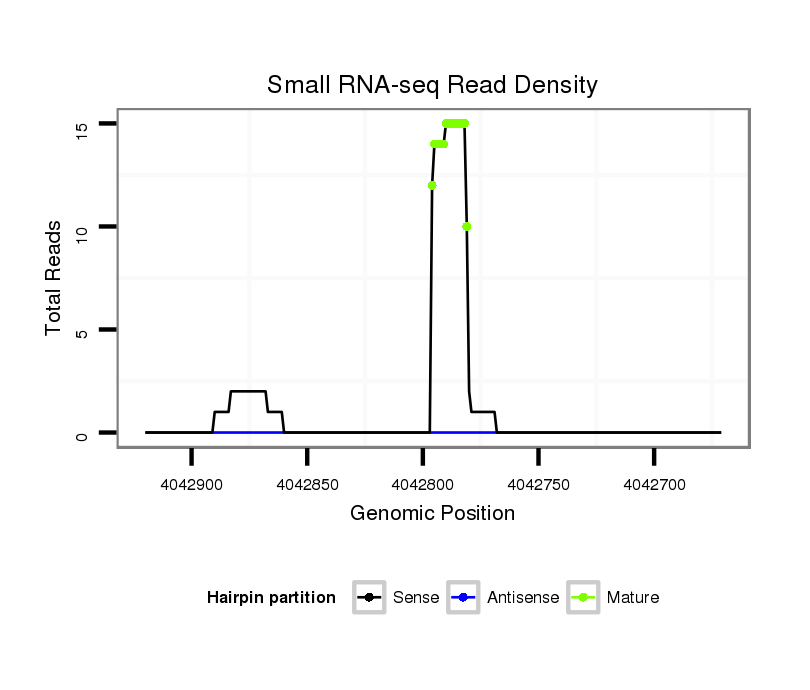
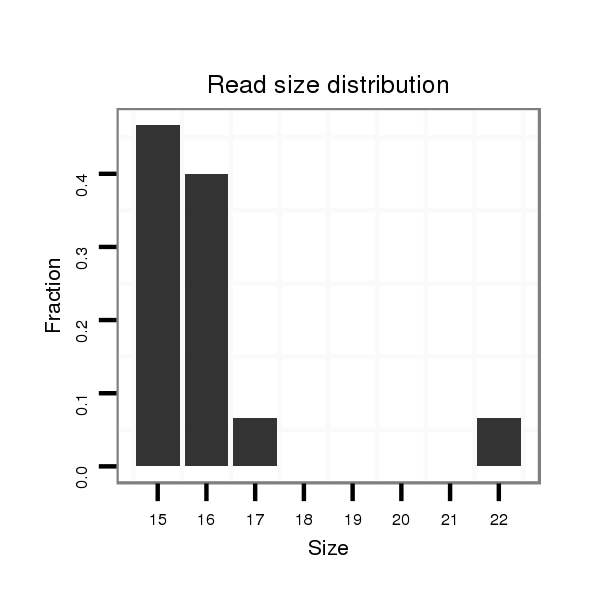
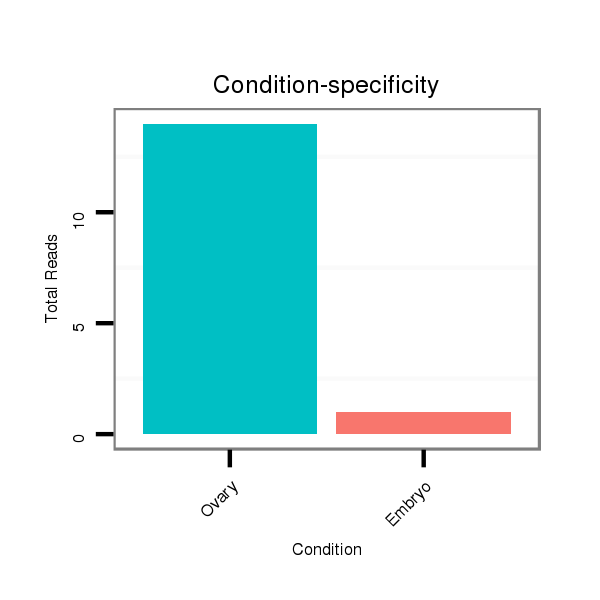
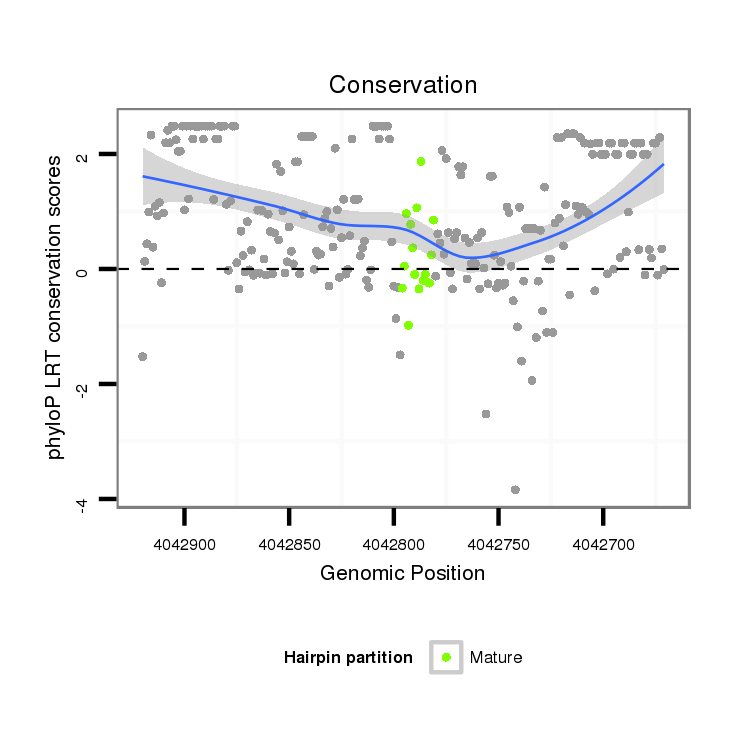
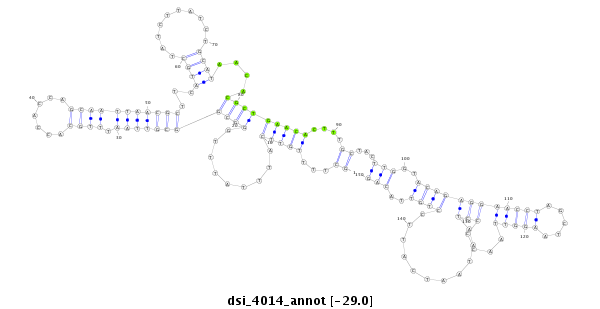
| ......................................................................................................................TTGCAGAACACGCTGAACACTTG............................................................................................................. |
23 |
3 |
1 |
39.00 |
39 |
39 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................TGCAGAACACGCTGAACACTTGA............................................................................................................ |
23 |
3 |
1 |
24.00 |
24 |
23 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................................................TTGCAGAACACGCTGAACACT............................................................................................................... |
21 |
2 |
1 |
20.00 |
20 |
20 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................TTGCAGAACACGCTGAACACTT.............................................................................................................. |
22 |
2 |
1 |
15.00 |
15 |
15 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................................................GCAGAACACGCTGAACACTTGA............................................................................................................ |
22 |
3 |
2 |
12.00 |
24 |
24 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................ATTGCAGAACACGCTGAACACTT.............................................................................................................. |
23 |
3 |
1 |
12.00 |
12 |
12 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................................................GCAGAACACGCTGAACACTTG............................................................................................................. |
21 |
2 |
1 |
11.00 |
11 |
11 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................TGCAGAACACGCTGAACACT............................................................................................................... |
20 |
1 |
1 |
10.00 |
10 |
10 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................................................GCAGAACACGCTGAACACT............................................................................................................... |
19 |
1 |
1 |
9.00 |
9 |
9 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................TGCAGAACACGCTGAACACTT.............................................................................................................. |
21 |
1 |
1 |
8.00 |
8 |
8 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................TTGCAGAACACGCTGAACA................................................................................................................. |
19 |
2 |
2 |
8.00 |
16 |
16 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................AACACGCTGAACACTTG............................................................................................................. |
17 |
1 |
2 |
7.50 |
15 |
15 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................ACACGCTGAACACTTG............................................................................................................. |
16 |
1 |
3 |
7.00 |
21 |
21 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................AATTGCAGAACACGCTGAACACTT.............................................................................................................. |
24 |
3 |
1 |
7.00 |
7 |
7 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................GAACACGCTGAACACT............................................................................................................... |
16 |
1 |
2 |
6.50 |
13 |
13 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................AACACGCTGAACACTT.............................................................................................................. |
16 |
0 |
1 |
6.00 |
6 |
6 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................ATTGCAGAACACGCTGAACACT............................................................................................................... |
22 |
3 |
2 |
5.50 |
11 |
11 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................AACACGCTGAACACT............................................................................................................... |
15 |
0 |
1 |
5.00 |
5 |
5 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................TGCAGAACACGCTGAACACTTG............................................................................................................. |
22 |
2 |
1 |
4.00 |
4 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................................................AGAACACGCTGAACACT............................................................................................................... |
17 |
1 |
1 |
4.00 |
4 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................AATTGCAGAACACGCTGAACACT............................................................................................................... |
23 |
3 |
2 |
3.00 |
6 |
6 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................CACGCTGAACACTTG............................................................................................................. |
15 |
1 |
9 |
3.00 |
27 |
27 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................................................AGAACACGCTGAACACTTG............................................................................................................. |
19 |
2 |
1 |
3.00 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................................................GCAGAACACGCTGAACACTT.............................................................................................................. |
20 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................GAACACGCTGAACACTT.............................................................................................................. |
17 |
1 |
2 |
2.00 |
4 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................ACACGCTGAACACTT.............................................................................................................. |
15 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................................................AGAACACGCTGAACACTT.............................................................................................................. |
18 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................................................................................CAGAACACGCTGAACACTT.............................................................................................................. |
19 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................TGCAGAACACGCTGAA................................................................................................................... |
16 |
1 |
4 |
1.25 |
5 |
5 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................TTGCAGAACACGCTGAA................................................................................................................... |
17 |
2 |
20 |
1.10 |
22 |
22 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................................................GCAGAACACGCTGAACACTTGT............................................................................................................ |
22 |
3 |
2 |
1.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................ATTGCATAACACGCTGAACACTTG............................................................................................................. |
24 |
3 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................TGCAGAACACGCTGAACA................................................................................................................. |
18 |
1 |
2 |
1.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................CTGCTTAGTACAGAGGAAC......................................................................................... |
19 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................................................GCAGAACACGCTGAACA................................................................................................................. |
17 |
1 |
3 |
1.00 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................................................................................................................................................TCATTCCTGTTACGGA................................................. |
16 |
1 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................................................GCAGAACACGCTGAATACTTG............................................................................................................. |
21 |
3 |
2 |
1.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................CTGAACACTTTGCTACTTGGTA.................................................................................................. |
22 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................CTGCAGAACACGCTGAACACTTGA............................................................................................................ |
24 |
3 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................TGCAGAACACGCTGAACAC................................................................................................................ |
19 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................AATTTGCATTTCATTTTCGGGCT..................................................................................................................................................................................................... |
23 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................ATTTCATTTTCGGGCTTTTGTTC.............................................................................................................................................................................................. |
23 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................TGCAGAACACGTTGAACACT............................................................................................................... |
20 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................CTGCGTAGTACAGAGGAACC........................................................................................ |
20 |
3 |
4 |
1.00 |
4 |
0 |
1 |
0 |
0 |
3 |
0 |
0 |
0 |
| ........................................................................................................................GCAGAACACGTTGAACACT............................................................................................................... |
19 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................................................GCAGAACACGCTGAACAC................................................................................................................ |
18 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................AACACGCTGAACACTTT............................................................................................................. |
17 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................GAACACGCTGAACACTTG............................................................................................................. |
18 |
2 |
12 |
0.75 |
9 |
9 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................................................................................CAGAACACGCTGAACACTTGA............................................................................................................ |
21 |
3 |
3 |
0.67 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................TGCAGAACACGCTGA.................................................................................................................... |
15 |
1 |
12 |
0.67 |
8 |
8 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................................................AGAACACGCTGAACAC................................................................................................................ |
16 |
1 |
3 |
0.67 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................AACACGCTGAACACTTGA............................................................................................................ |
18 |
2 |
9 |
0.67 |
6 |
6 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................GAACACGCTGAACAC................................................................................................................ |
15 |
1 |
8 |
0.63 |
5 |
5 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................ACACGCTGAACACTTGA............................................................................................................ |
17 |
2 |
20 |
0.60 |
12 |
12 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................TGCGTAGTACAGAGGAACC........................................................................................ |
19 |
3 |
14 |
0.50 |
7 |
0 |
3 |
0 |
0 |
1 |
0 |
3 |
0 |
| .......................................................................................................................TGCAGAACACGTTGAACACTTG............................................................................................................. |
22 |
3 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................................................AGAGTTGTAGCAAATAACC................................. |
19 |
2 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| ......................................................................................................................TTCCAGAACACGCTGAACACTT.............................................................................................................. |
22 |
3 |
2 |
0.50 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................................................ACAGAACACGCTGAACACTTG............................................................................................................. |
21 |
3 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................AAACACGCTGAACACTT.............................................................................................................. |
17 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................TGCAGAACACGCTGAAC.................................................................................................................. |
17 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................................................TCAGAACACGCTGAACACTTG............................................................................................................. |
21 |
3 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................TTGCAGAACACGCTGAAC.................................................................................................................. |
18 |
2 |
7 |
0.43 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................................................................................CAGAACACGCTGAACACTTGT............................................................................................................ |
21 |
3 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................AATTGCAGAACACGCTGAACAC................................................................................................................ |
22 |
3 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................................................GCAGAACACGCTGAA................................................................................................................... |
15 |
1 |
16 |
0.25 |
4 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................ATTGCAGAACACGCTGAACAC................................................................................................................ |
21 |
3 |
8 |
0.25 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................................................AGAACACGCTGAACACTTGA............................................................................................................ |
20 |
3 |
10 |
0.20 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................................................................................................................TTCATTCCTGTTACGGA................................................. |
17 |
2 |
7 |
0.14 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .....TTTTAGGTCATTGCA...................................................................................................................................................................................................................................... |
15 |
1 |
14 |
0.14 |
2 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
1 |
| .........................................................................................................................CATACCACGCTGAGCAC................................................................................................................ |
17 |
2 |
17 |
0.06 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |