ID:dsi_3838 |
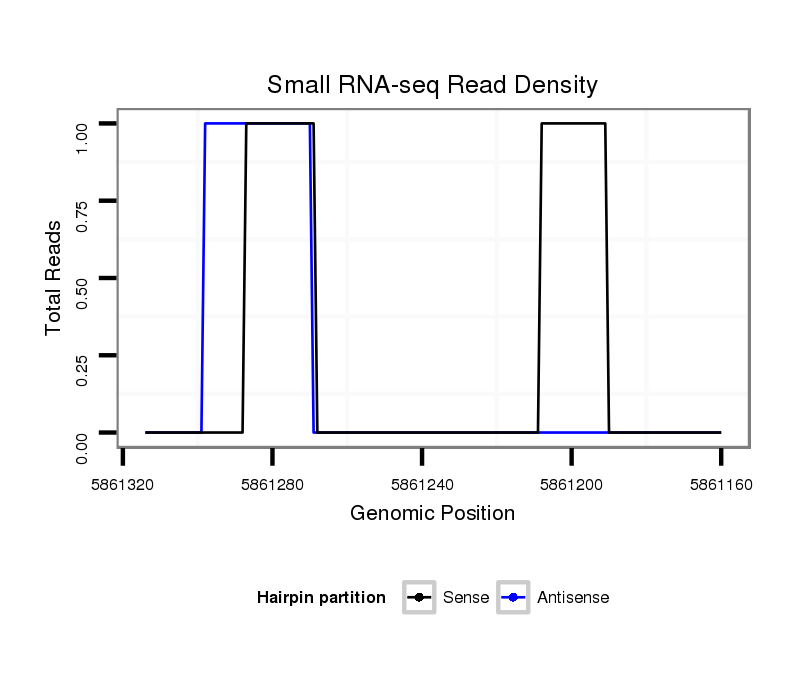
Coordinate:2r:5861210-5861264 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
exon [2r_5860687_5861209_-]; CDS [2r_5860687_5861209_-]; CDS [2r_5861265_5862438_-]; exon [2r_5861265_5862438_-]; intron [2r_5861210_5861264_-]
No Repeatable elements found
| ##################################################-------------------------------------------------------################################################## ATTGCCCAGCTCTCAGAGCTAGTTTCCAGCTTTGAGTTCGATGCACACAAGTAAGTGATCAGAACATAAAAACCGGTTTTATATAACGTTAATTTTATCCTTAAGGGACGAGCTGCAGATCGGCGGTATATTCATACGCATCTACAATGATATGC **************************************************.((((.(((...(((((((((.....)))))).....)))......)))))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902009 testis |
M024 male body |
M025 embryo |
M053 female body |
O002 Head |
SRR618934 dsim w501 ovaries |
SRR553485 Chicharo_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................GAGCTGCAGATCGGCGGTATATTAT..................... | 25 | 2 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................AGGACGAGCTGCAGATCGGCGGTATA......................... | 26 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................GTTCGATGCACACAAGGACG.................................................................................................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................AGCTTTGAGTTCGATGCAC............................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................GACGAGCTGCAGATCGGC............................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................ACGAGCTGCAGATCGGCGGGT........................... | 21 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............AGAGCTAGTTTCAAGCT............................................................................................................................ | 17 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................TCAGAACATAAAAACT................................................................................. | 16 | 1 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TAACGGGTCGAGAGTCTCGATCAAAGGTCGAAACTCAAGCTACGTGTGTTCATTCACTAGTCTTGTATTTTTGGCCAAAATATATTGCAATTAAAATAGGAATTCCCTGCTCGACGTCTAGCCGCCATATAAGTATGCGTAGATGTTACTATACG
**************************************************.((((.(((...(((((((((.....)))))).....)))......)))))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
M023 head |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR902008 ovaries |
M053 female body |
SRR553485 Chicharo_3 day-old ovaries |
M025 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..ACGGGTCGGGAGTCTC......................................................................................................................................... | 16 | 1 | 3 | 6.33 | 19 | 0 | 14 | 4 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..ACGGGTCGGGAGTCTCGGTTA.................................................................................................................................... | 21 | 3 | 3 | 6.00 | 18 | 11 | 1 | 1 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| .CACGGGTCGGGAGTCTCGGT...................................................................................................................................... | 20 | 3 | 11 | 1.73 | 19 | 13 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | 0 |
| ..ACGGGTCGGGAGTCTCGGTT..................................................................................................................................... | 20 | 3 | 15 | 1.53 | 23 | 18 | 1 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................CGATCACAGGGCGAATCTCAA..................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................TCGATCAAAGGTCGAAACTCAAGCTACGT.............................................................................................................. | 29 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .AACGGGTCGGGAGTCTCGGTTA.................................................................................................................................... | 22 | 3 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GCACGGGTCGGGAGTCTCGAT...................................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...CGGGTCGGGAGTCTC......................................................................................................................................... | 15 | 1 | 7 | 0.86 | 6 | 0 | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GCACGGGTCGGGAGTCTC......................................................................................................................................... | 18 | 3 | 20 | 0.85 | 17 | 7 | 4 | 1 | 0 | 2 | 0 | 1 | 0 | 0 | 2 |
| ...CGGGTCGGGAGTCTCGGT...................................................................................................................................... | 18 | 2 | 6 | 0.67 | 4 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..ACGGGTCGGGAGTCT.......................................................................................................................................... | 15 | 1 | 7 | 0.57 | 4 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| .CACGGGTCGGGAGTCTCGATTA.................................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..ACGGGTCGGGAGTCTCGGT...................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................ACGTATGCGTAGATGATATTAT... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................CTGGTCGATGTCTAGCAGCC............................. | 20 | 3 | 9 | 0.44 | 4 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| .CACGGGTCGGGAGTCTC......................................................................................................................................... | 17 | 2 | 8 | 0.38 | 3 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................TCACGCTAGGTGTGTT......................................................................................................... | 16 | 2 | 20 | 0.35 | 7 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 |
| .CACGGGTCGGGAGTCTCG........................................................................................................................................ | 18 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................TTAAAATAGGAATTC.................................................. | 15 | 0 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................GCTCGATGTCTAGCAGCC............................. | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....GGGTCGGGAGTCTCG........................................................................................................................................ | 15 | 1 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GCACGGGTCGGGAGTCTCG........................................................................................................................................ | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .CACGGGTCGGGAGTCT.......................................................................................................................................... | 16 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...CGGGTCGTGAGTCTC......................................................................................................................................... | 15 | 1 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................CTTGTCGATGTCTAGCCGCC............................. | 20 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....GGGTCGGGAGTCTCGGT...................................................................................................................................... | 17 | 2 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................AAAAAAGGAACTCCCTGAT............................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TGTTCGATGTCTAGCAGCC............................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
Generated: 04/24/2015 at 01:30 AM