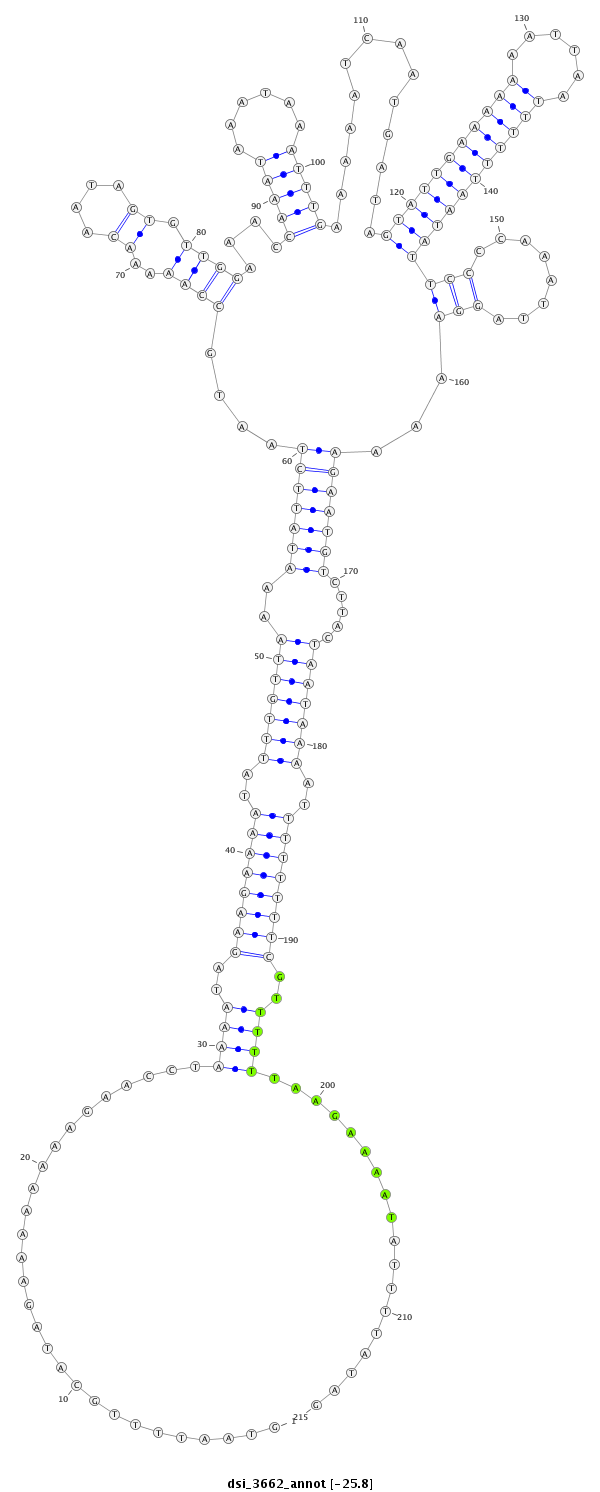
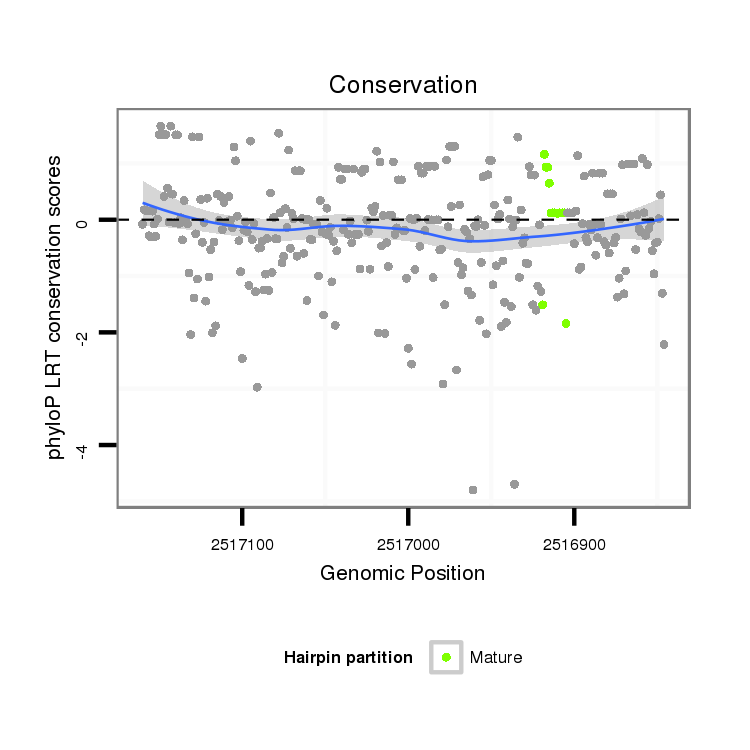
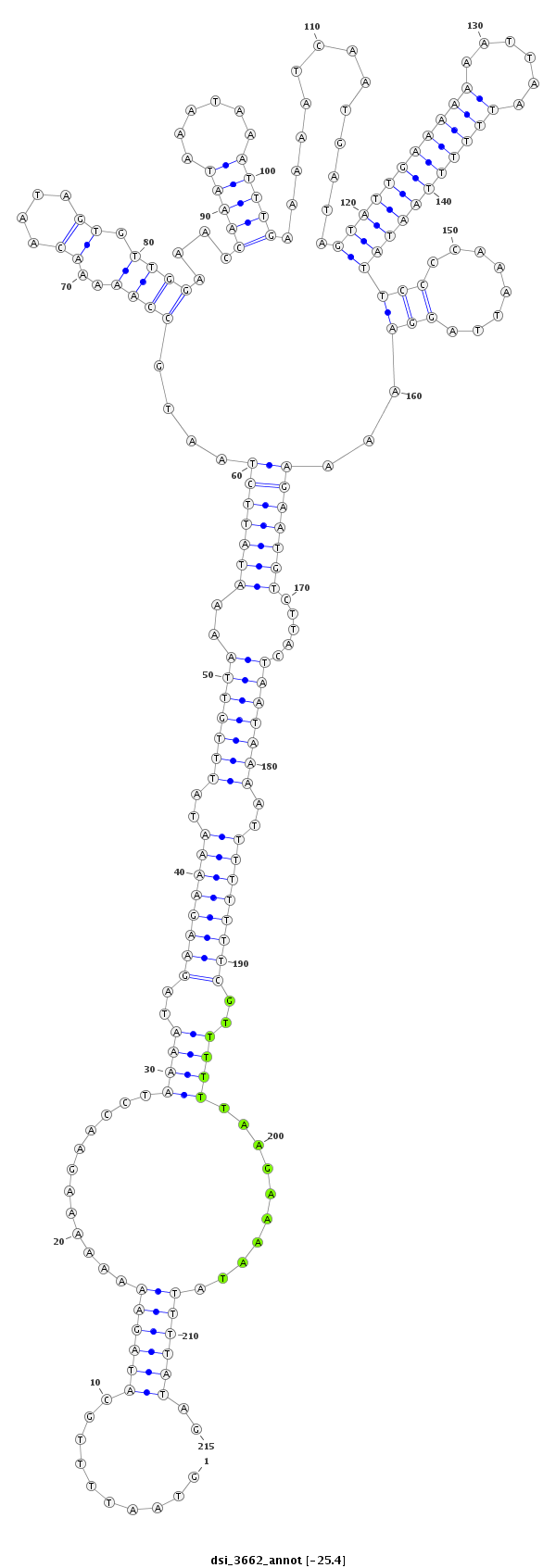
ID:dsi_3662 |
Coordinate:3l:2516896-2517110 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -25.4 | -25.4 | -25.4 |
 |
 |
 |
five_prime_UTR [3l_2517111_2517265_-]; exon [3l_2517111_2517265_-]; five_prime_UTR [3l_2516510_2516895_-]; exon [3l_2516340_2516895_-]; intron [3l_2516896_2517110_-]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| mature | star |
| ##################################################-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TTCGCGGAGGCACTGAGAGAAAACTAGTGCTCGTATCGCTAGTAATTTCGGTAATTTTGCATAGAAAAAAAAGAACCTAAAATAGAAGAAAATATTTGTTAAAATATTCTAATGCCAAAAACAATAGTGTTGGAAACCAAATAAATAAATTTGAAAAATCAATGATAGTATTGAAAAAAATTAATTTTTTAATATTCCCCAAATTAGGAAAAAGAATGTCTTACTAATAAAATTTTTTTTCGTTTTTTAAGAAAATATTTTATAGACGTGATCGCAATTATTTATGGTTTCCAAAATTTTAAAATTAAGTTTTTC **************************************************............................((((..((((((((..(((((((..(((((((....((((..((....)).))))....(((((......)))))..............(((((((((((......)))))))))))(((........)))...))))))).....)))))))..))))))))..))))..................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
M053 female body |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
M024 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................TCGTATCGCTAGTAATC............................................................................................................................................................................................................................................................................ | 17 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| TTCGCGGAGGCACTGAGAGAAAACTA................................................................................................................................................................................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................TCGCTAGTAATTTCGG........................................................................................................................................................................................................................................................................ | 16 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................TTTCTGGTTTCCAAAATT................. | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................TTGTTAGAATATTCGAATCCCAA..................................................................................................................................................................................................... | 23 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................ATAAAATTCTTTTTCGGTTTT.................................................................... | 21 | 2 | 4 | 0.50 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................ATCGCTAGTAATTGCGG........................................................................................................................................................................................................................................................................ | 17 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................TGTAATTTTGCAGAGAAAAAAAA................................................................................................................................................................................................................................................... | 23 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................GAATAAAATTCTTTTTCGGTT...................................................................... | 21 | 3 | 9 | 0.33 | 3 | 0 | 0 | 0 | 0 | 3 | 0 | 0 |
| ...................................TCGCTAGTAATCTCGG........................................................................................................................................................................................................................................................................ | 16 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TAATCTTGCATCGAAAAAA..................................................................................................................................................................................................................................................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................AATTTTGCATCAGAAAAAAAGA................................................................................................................................................................................................................................................. | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................................................................CCGAATGAGGAAAACGAATG................................................................................................. | 20 | 3 | 9 | 0.22 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| .................................................................................................................................................................................................................................................GTTTTTTAAGAAAAT........................................................... | 15 | 0 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................TCGCTAGTAATCTCG......................................................................................................................................................................................................................................................................... | 15 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................AAACTAGTGCTCTTA........................................................................................................................................................................................................................................................................................ | 15 | 1 | 13 | 0.15 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................AATTTGGAGAAAGAATGGCT.............................................................................................. | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................TAGTGCTCGTCTCGGT................................................................................................................................................................................................................................................................................... | 16 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ...............................................................................................................................................................CAATGATTGTACTTAAAAAAA....................................................................................................................................... | 21 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| TTCTCAGAGGCACTGAAAGA....................................................................................................................................................................................................................................................................................................... | 20 | 3 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................AATCGCTAGTAATTCCGA........................................................................................................................................................................................................................................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................TTCCCCAAATTGGGAGGAA...................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................AAATGATAGTATTGA............................................................................................................................................. | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................TCGCTAGTAATCTCTG........................................................................................................................................................................................................................................................................ | 16 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................TTAGTATTGAAAAAAA....................................................................................................................................... | 16 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
|
AAGCGCCTCCGTGACTCTCTTTTGATCACGAGCATAGCGATCATTAAAGCCATTAAAACGTATCTTTTTTTTCTTGGATTTTATCTTCTTTTATAAACAATTTTATAAGATTACGGTTTTTGTTATCACAACCTTTGGTTTATTTATTTAAACTTTTTAGTTACTATCATAACTTTTTTTAATTAAAAAATTATAAGGGGTTTAATCCTTTTTCTTACAGAATGATTATTTTAAAAAAAAGCAAAAAATTCTTTTATAAAATATCTGCACTAGCGTTAATAAATACCAAAGGTTTTAAAATTTTAATTCAAAAAG
**************************************************............................((((..((((((((..(((((((..(((((((....((((..((....)).))))....(((((......)))))..............(((((((((((......)))))))))))(((........)))...))))))).....)))))))..))))))))..))))..................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
GSM343915 embryo |
M024 male body |
M025 embryo |
SRR553486 Makindu_3 day-old ovaries |
M053 female body |
M023 head |
SRR553488 RT_0-2 hours eggs |
SRR902009 testis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................................................................CCTTCTTGTATAAAATATCTGCA.............................................. | 23 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................ACCAAAGGTTTCTAAATTAT........... | 20 | 3 | 20 | 1.00 | 20 | 0 | 0 | 8 | 6 | 0 | 3 | 2 | 0 | 1 |
| ..................................................................................................................GGTTTTTGTGATCACAAGCT..................................................................................................................................................................................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................AGGATAGCGCTCATT.............................................................................................................................................................................................................................................................................. | 15 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........GAGACTCTCTGTTGAT................................................................................................................................................................................................................................................................................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................ACCAAAGGTTTCTAAATT............. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................TGTTTTTAGTTACTATCA.................................................................................................................................................. | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................ACTGTCTGCACTAGCG........................................ | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................TTTTCTTAAAGAATG........................................................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 3l:2516846-2517160 - | dsi_3662 | TTCGCGGA--GGCACTGAGAGAAAA-CTAGTGCTCGT-----------------------AT---CGCTA------------------------------------------------GTAATTTCGGTAATTTTGCATAGA---A-AAAAAAG--------------------------------------------------------------------------------------------------------------------------------------------------------------------AACCTAAAAT----AGA-----AGA-------------------AA-----------------------------------------------------------------------------ATATTTGTTA----------------------AAATAT------------------------------TCTAA-----------------------------------------------------------------TGCCAAAAACAATAGTGTTGGAAAC-CAAATA---AATAAATTTGAAAAATCAATGATAGTATTGAAAAAAA-TTAA--TTTTTTAA----------------TATTCC--------------------------------------------------------------------CCAAATTA-----------------------------------------------------------------------------------------------GGAAAA------------------AGAATGTCTTACTAA-TA---------AAA-------------------------TTTTTTTTCGT-------------------------------------------------------------------------------TTTTTAAGAAAATATTTTATAGACGTGATCGCAATTATTTATGG---------------------------TTTCCAAAATTTTA---AAAT--------------------TAAGTTTTTC |
| droSec2 | scaffold_11602:155-431 - | TTCGCGGA--GGCACTGAGAGAAAA-CTATCGCTCGA-----------------------AT---CGCTG------------------------------------------------GTAA-TTCGGTAATTTTGCATAGC---A-AAGCGAG--A---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGA-------------------AA-----------------------------------------------------------------------------ATATTGGTTA----------------------AAATAT------------------------------TCGAA-----------------------------------------------------------------TGTCCAAAACAATAGTGTTGGAAAT-CAAGTG---AATCAATTTGAAAAATCCAGGATAGTATTGAAAAAAAATCAATTTTTTTGAA----------------TATTCC--------------------------------------------------------------------CAAAATTA-----------------------------------------------------------------------------------------------GAAAAT------------------AGATTGTTTAAAAAA-AA---------AAA---------------------------TATACTCGT-------------------------------------------------------------------------------TTTTTAAGAAAAGATTTTATAGA---------------------------------------------------CAAACATTTTT---AAAT--------------------TA--TTTTTC | |
| dm3 | chr3L:2629651-2629962 - | TTCGCGGA--GGCACTGAGAGAAAA-CTATCGCTCGA-----------------------AT---CGCTA------------------------------------------------GTAA-TTCGGTAATTTTGCTTAGC---A-AAGCGAG--------------------------------------------------------------------------------------------------------------------------------------------------------------------GACCAGAAAT----AGG---A-AGA-------------------AA-----------------------------------------------------------------------------ATATTTGTTA----------------------AAATAAAAAA------------------AATAGTTGTCTAA-----------------------------------------------------------------TGCCCAAAACAATAGTGTTGGAAAA-CAAATG---AACAAATTTGTAAAATCCAAGATAGTATTGAAGAAAAAAAGAATTTTTTAAA----------------TATTCC--------------------------------------------------------------------CT----------------------------------------------------------------------------------------TTGGAATTAGATAGGAAAT------------------AGATTGGATTACTAA-AAATTATATTAAAT---------------------------TATATTCGT-------------------------------------------------------------------------------TTT---------------ATAG-----------------------------ATACG---------------TTTCCAAAATTTTT---AAAT--------------------TAAGTTTTTC | |
| droEre2 | scaffold_4784:2648716-2649001 - | TTCGCGGG--GGCACTGAGAGAAAA-CTGTCGCTCGA-----------------------AT---CGTTA------------------------------------------------GTAG-TTCGCTACTTTTGCATTGC---A-AAAAAG-ATGGGA-GC----------------------------------------------------------------------------------TCAATTATA------------------------------------------------------AGACTTTGTGGCACTGAAGT----AGA-----AGA-------------------GT-----------------------------------------------------------------------------GTATTTCTAA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATATTTAAGTGAACAACTGAATTGGAAGAATATATTTCTAAATATTTAAATGACGAACTGAGTTCGAGGAAAATATTTCTAAATATTTAAATGACCA-------------------ACT----GAGTTCGAAGAAA---ATATTTCCAAA------------------------------------------------------------------------------------TA-----------------------------------------------------------------------------------------------------------------------------TTT---AAGTGACCAACTGA---ATTGGAAGAATATATTT | |
| droYak3 | 3L:9306655-9306919 + | TTCGCGGA--GGCACTGAGAGAAAA-CAGTCGGTCGA-----------------------AT---CGTTA------------------------------------------------GGAA-TTCGGTAATTTTGATTAGC---A-AAACAGG--GAGAAGTTTTAG-----------------------------------------------------------------------------CCTATAGTA------------------------------------------------------AAAACTTGTGGAACTAAAAT----AGA---A-AAA-------------------GA-----------------------------------------------------------------------------ATATTTGTAA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTTTAAAATGCCCAACTGAAATAAAAGAAAATATTTATAAATTATAAAGTGACTAACTGAAATAGAAGAAAGTATTACTTAATATTTAAATGGCAA-------------------ACT----GAAATAGAAGCAA---AAATTTCGAAA------------------------------------------------------------------------------------TA-----------------------------------------------------------------------------------------------------------------------------TTT---AAAT------------------------------ | |
| droEug1 | scf7180000409466:356346-356686 - | TTCGCGAA--GGCACTGAGAGAAAA--TATCAGTGGT-----------------------AT---TGGTA------------------------------------------------AAGT-TTCGGTAACTTTCGCTTGTTGTA-AA-AAAA--GAGAG-C-TTTGCTA--------------------------------------------------------------------------GCTA----TAATTTAAAGTTAAAAAAGATTTATAA----AACTCAATTCAA-------------------ATATTTTTTACTT-----TG-----AAA-------------------AACTGTTTAACCTTTTTAGTCTTATGGT-----------------------------------------------------------------------------------------------------------------------CTTAA-TTTGCAA---------------------------------------------------------GACC------AAAACTAAACGAAATTTAAATTAGAAATTTGTTTTAAAAAATAGAACAAATATTAAAAAACG-TTTA--TTTTTAAA----------------TGAGTT--------------------------------------------------------------------TTAAATCA-------------------------------------------------------------------------------------------AA-------------------TCGAATAAGATCAAATTTCTGAA------------------------------------------------------------------------------------TA--TTTTTTGTTTAA-------------------------------------------------------------------------------------------------------------------------------CTTTTTTATTATAATTT-CTTTCC | |
| droBia1 | scf7180000302428:9407932-9408341 - | TTCGCGGG--GGCACTGAGAGAAAA--TATCTGTGAA-----------------------AT---CGCTT------------------------------------------------GCCT-TTCGGTAA--------AGC---A-AA-CAAG--GAGAA-C-TGAGCATCTATTCAAACTGTGTAA--AAACTTTAATATTTTAAAATATTTTAAAAATCAGTGCTTGCAAAGTGGAATACTAATTATGGCA------------------------------------------------------TAAAACTATTTTTTTATTTTTATTATAC-AATAAAAACTGCTTAGTGGCAAACCAA-----------------------------------------------------------------------------ATATTTATAG----------------------TGTATATTAAAAGCTTCAATTTCTTTTTTGTAGAATTCCTA--T-----------------------------------AATAAAAAACA----------------------------------------TTAAAATAA------------------------------------------TA--TTTTTACA----------------AGTGTC--------------------------------------------------------------------CCAAAA-------------------------------------------------------------------------------------------------GAA----------------------------------------------------------------------------------------------------------------------AATTGTGTTTTTTGTTTAATTTGAAATAATTA---------------------------------TTTTTACAAGATCTTAATACTATAAGCCTTAAAGGAATGACTCTTATTTTCATAA--------------------------CTTAAAATC---------TAAAAT | |
| droTak1 | scf7180000415857:426184-426419 - | AAATAT--------------------------------------------------------------------------------TTAAAAATTAAAAATGTATCATAGCATAAATAATAATCAAGGCAAC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACTTATACTTTTATTAGT-----AAA-------------------AA-----------------------------------------------------------------------------ATCTTTATCA----------------------TGATTATTTA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AATTGCTTTTTCAAATATCCCAAAAATTAGT----------------------------------------------------------------------------------------------------------------------------TTCATAAA-CT---------GGA-------------------------TTTTTTTTAAT------ACACATTTATTACAAATGTGGCAAAAACCGTGATTTTT-TTTAATAGTTAATAGTTA---------------------------------ATTA--------------------------------------------------CATATAAAATCTTA---AAA----------------------ATGTCTTAA | |
| droEle1 | scf7180000491249:2750937-2750959 + | TTCGAGGA--GACACTGAAAGAAAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000758401:32919-33387 + | TTCGCCGA--GACACTGAGAGAAAA--TATCGGTAAC-----------------------AT---TGCTT------------------------------------------------ACAG-TTCGATGAGTTTTCAAATC---A-GTAAAG-AGTGTA-GT----------------------------------------------------------------------------------TCTATCGCT------------------------------------------------------GGTTTAAATTAAACTTAAAT----GGA-----AAA-------------------CACTTTTTAACACCCTAAATTTTAAGATTTTAAAATTTTACTGAAATATAATTTTAGAAGTTCAGATCTTTAGTTAAAGATTTTTAACACATATTATTAATATACAAGTGAAAATTTAAAA------------------ATCTGCTGT--------------------------------------------------------------------------------------------CT-CTAATA---AGTAATTTTGAAAAGTGTAAAGTAA--CTGATTATAAATAAAACTTTTTTGA----------------GATTACATTAATTTGGAAAGTGTTAAAGAAACTTTACAAATTATAATTTAGA--------------------------------------------------------------------------------------------------------------------------------AAT------------------TGTTGGTATTACTTA-AAATAAACTAGAGTTAAATTTAAAAATAAGTTATACATAAT------GCTTTACCAC--------------------------------------------------------AAATGTTGTATAA--------------------------------------------TAT---------------------------TTTGCAAAATTCATAAGAAAT--------------------TGTATTTTTA | |
| droFic1 | scf7180000454105:956903-956924 - | TTCGCGGA--AGCACTGGGAGAAA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droKik1 | scf7180000302441:1077175-1077625 + | A-CGCGGCAACGCACAGAAAGC--C-ATATCTCTGAAAAAATATTTAAGAAAAAGGTTGGATAATCTGTG------------------------------------------------AAAT-TTTAGAAACTTGGTCAAAT---ATTAGTGAA-----A-------------------ACTCTAAAAAAAAAGTATAG-----------------------------------------------TAA----TTATCTGAAATATAAAAAAAAATGTAGTACAAAATTAATACAAATAATAATTTGGATTGGCTAATTTTGTAATTTTATAAAAAGAAACAATTCTAGTTTGTG--------------------------------GAT-----------------------------------------------------------------------------------------------------------------------TTTAA-TATTGAAAATGTGATAATTTTAGGAAAATTTATTATTAAAAAAAAACATTACACATATGATAAGTGAATTA---------------------------------------------AATAATAGTACTGAAAAAATAA-TA--ATCTTAAAAAAGTGTGAAAAATTTT-ATCT--------------------------------------------------------------------CT----------------------------------------------------------------------------------------TTAGGTTTTTACAACAAA------------------AAA--AGAATTC-GAAAAAATCGAATTGGAT-------------------------A-TTTACCTGT-------------------------------------------------------------------------------TTT---------------TTTC-----------------------------AAATA---------------A--------------------------ATTTAAAATTAAAATACAGTTTTT | |
| droAna3 | scaffold_13337:9451709-9452019 + | TTCGTAAA--TACACAGAAAACAAA-ACAACACTTGG-----------------------AT---TATAA------------------------------------------------GTTA-ATTGGTAGCT--GCTTAAG---A-GTAAA----GGAT-TT----------------------------------------------------------------------------------TTTGTTATT------------------------------------------------------AAATCCTGTGTCTTTAATTTTAAT------------------------------------------------------------------------AAAG-----------------------------------------------------------------------------------TTTAAGTTTGTTTAGTGGTTTAATAT-----------------------------------------------------------------TACAAATAAATGTAAATGAAAT-AAAAT------------------------------------------------GTTTTTGT----------------GTTTA----AACTTTAAAAACATTAATGTATTTTTTCAAAACAGTATCTGGG--------------------------------------------------------------------------------------------------------------------------------AACACTTCTAAAGTTCTAAAAATATTAAAATTCCAAA-------------------------A----------------------------------------------------------TATATTGTTAATTTAAT----------TA----------------------------------TTGTTCAAACTTTTATTAC-----------------------------------------------T---ATTT--------------------TATGTTTTGA | |
| droBip1 | scf7180000396544:262667-262968 + | GTTTTAAA--TACACAGAAAATAATAAAAAAAATCGG-----------------------AA---TATAAGCCAATCGGTAGCTGCTTTAAAATTAAAGATTTATTATTTTT---------------------------------------------------------------------------------------------------------------------------------------------G------------------------------------------------------AAATACTGGGTCTTTAATTTTATT------------------------------------------------------------------------TAAG-----------------------------------------------------------------------------------ATTACGTTTTTTGAGTGGTTCTGTAG-----------------------------------AATAAAAAAAA----------------------------------------TTAAGATTAGAATTCTGCAT----------------------------------------------------------------------------------------------------TATAATTTATAAATGTTTTT--------------------------------------------------------------GTTTAAATAAAAAAA-------------------------------------------------------------------GGATTTCCGAA-AAAGGATTTGAAGTTAAA---------------------GT------TCTCTTACGCCA------------------------------------------------------AAACTTTCTGTAACGTTT---------------------------------------------------------------------TTTTCAAAATATAT--------------GTTTTTATTTCAAAAC-TATTCT | |
| dp5 | XR_group3a:1310321-1310328 + | TCCGTAGA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_23:1341862-1341869 + | TCCGTAGA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/18/2015 at 09:44 AM