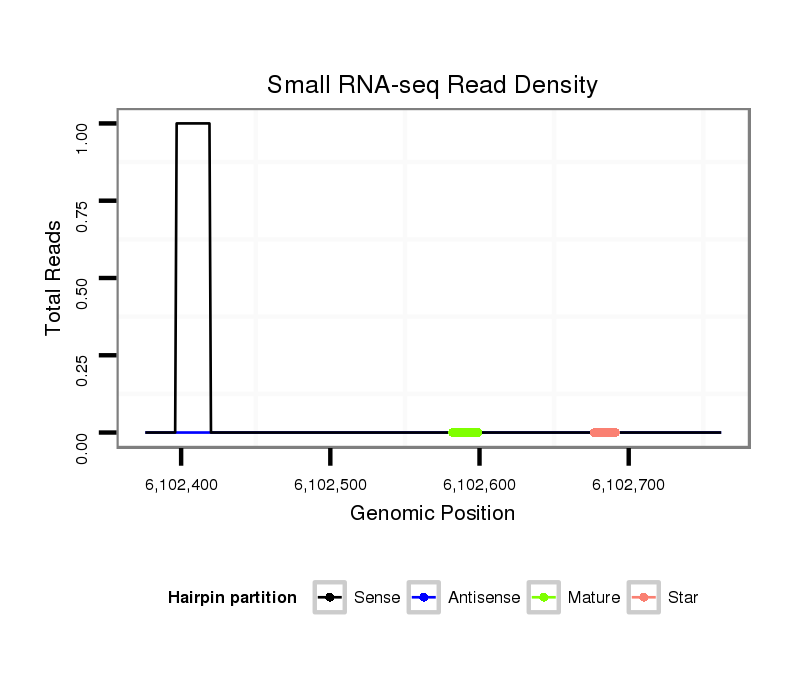
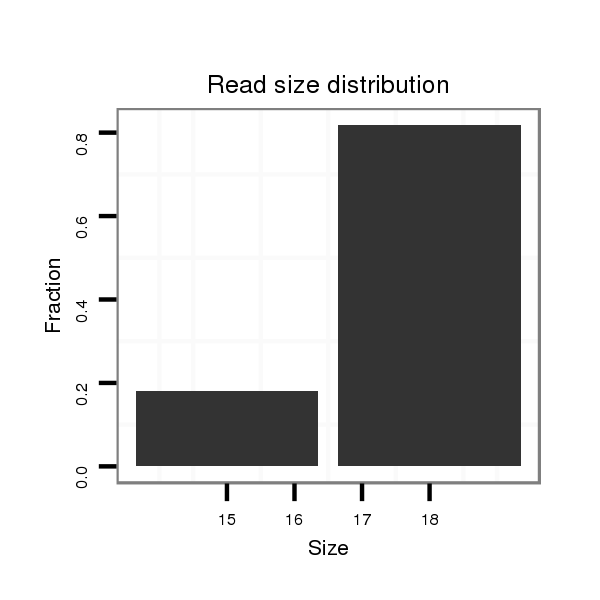
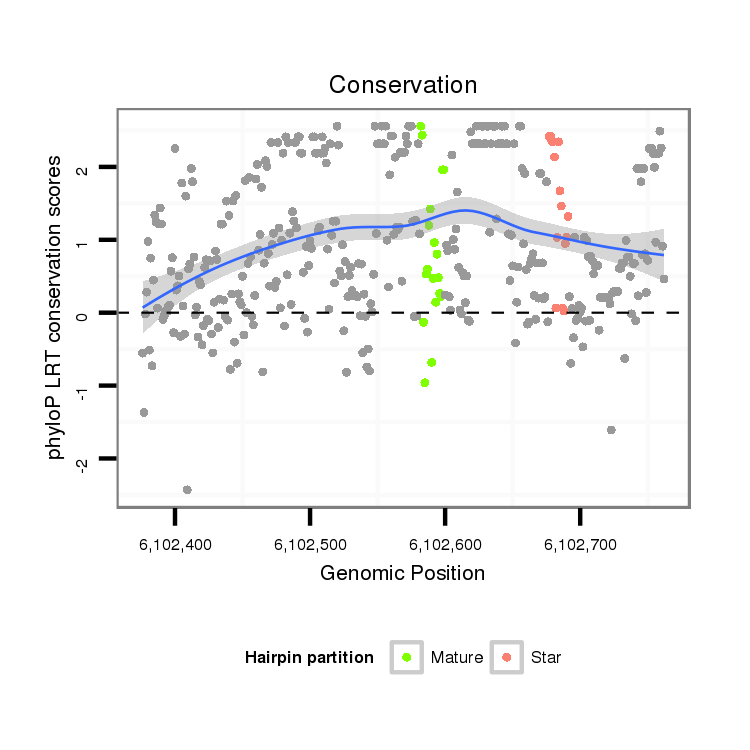
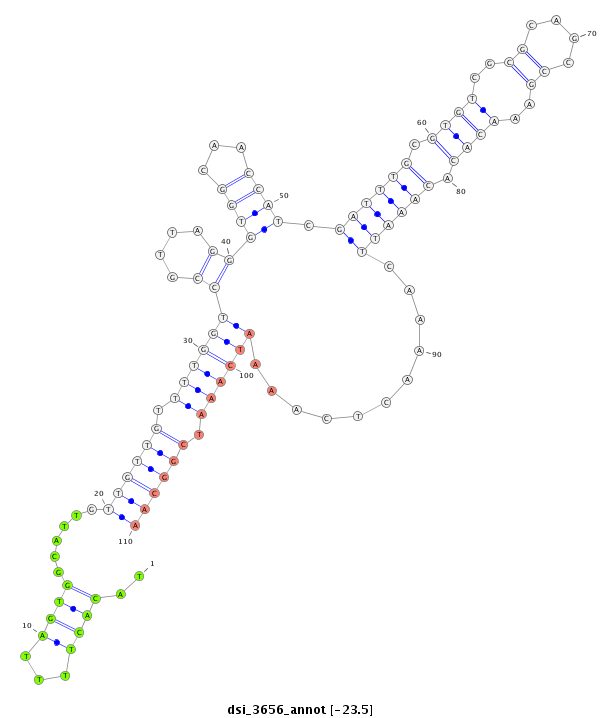
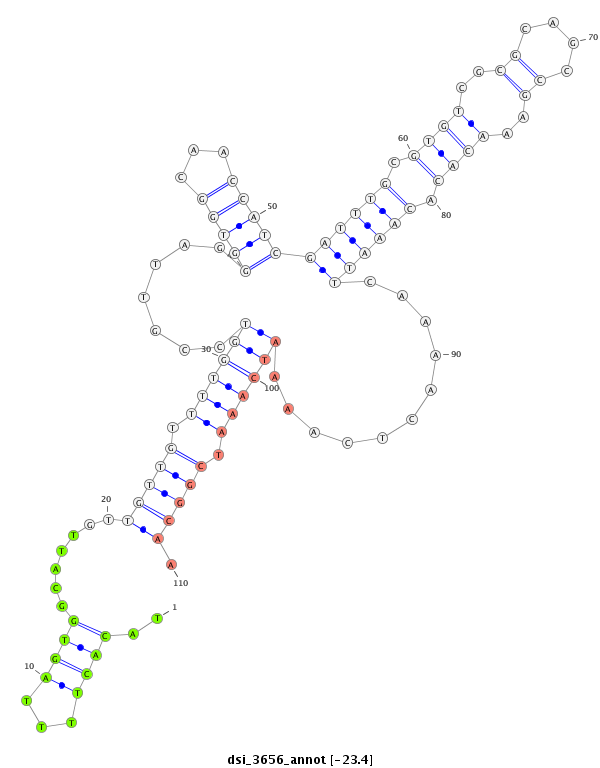
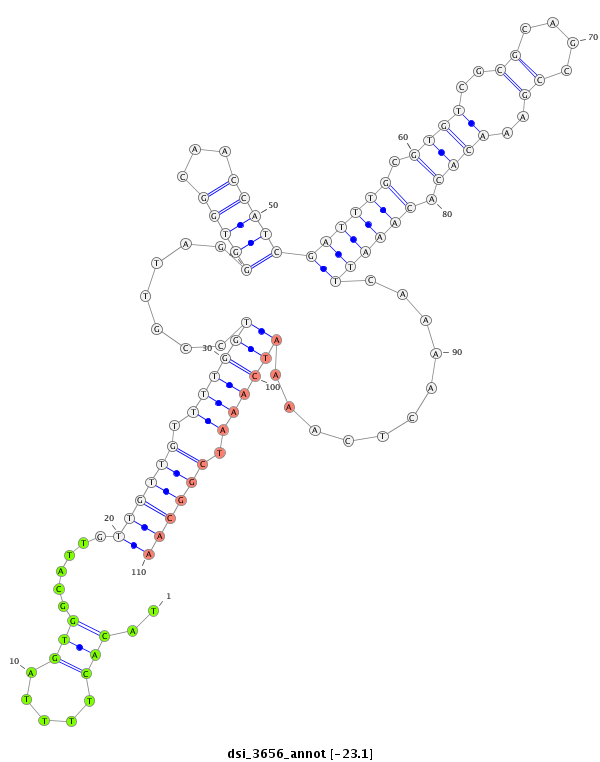
| droSim2 |
x:6102376-6102762 + |
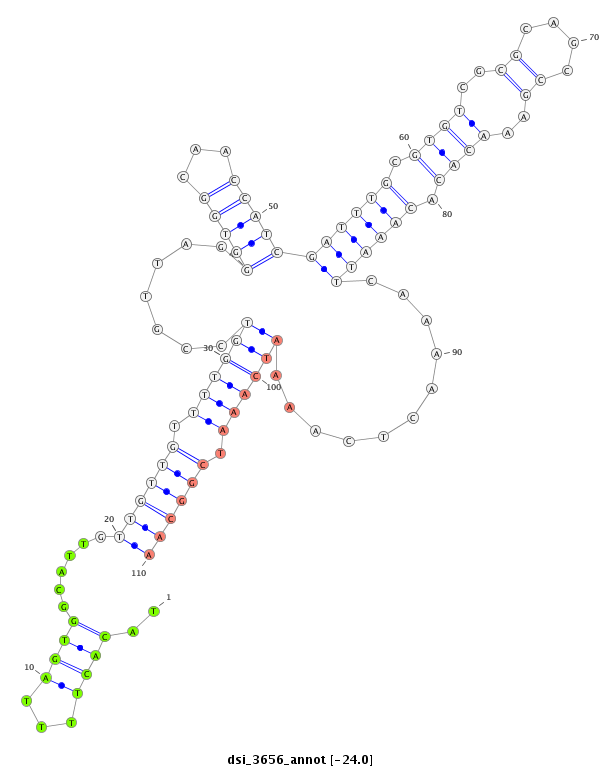
dsi_3656 |
GATGTTTAGTGA---T---------------TTC----C-----AGCTGTGT------GT-----------GGTGCTGT--------------------GTG----AA-TATTGAGAAGG-TG-------------------------------------A------------------AACATTG-----GTGTTTT--T-TGGCATCGCTGTTGTTTT--T--TCG-----------------------------TAGCACACTTACCGCTAT-AATAATAACGTGCTTCCTTCCTTGTC--------------------------------------------TGG---CCGAT------------------TT-TTCGGTGCC---G-----------------------------------------------CCACCTCTGCCGT-------------CG--CGTCGCGTGCAG-AAAA------------------------------AGTAAGCGCAGAATTAAACGTACACTT-TTAGT------G----------------------------------------------------------------------------G------------------------------------------------------CATTGTTGTT---G-TTT----------------TGGTCCG------TTAGGGTGGCAACCATCGATTTGCGTGTCGCGCAGCCGA--AA--------------------------C-ACACA--AATTCAAA------ACTCAAAATCAAATCGGCAAAAAGAGTAGACATA--ATAAA--AAAGTATATATACCTATA--------TATAAATA----CAGGGA--AACAAAAGAAATCTAATGCC-------- |
| droSec2 |
scaffold_4:304408-304801 - |
|
GGTGTTTAGTGA---T---------------TTC----C-----AGCTGTGT------GT-----------GGTGCTGT--------------------GTG----AA-TATTGAGAAGG-TG--------------------------------------------------------AACATTG-----GTGTTTT--T-TGGCATCGCTGTTGTTTT--T-TTCG-----------------------------TAGCACACTTACCGCTAT-AATAATAACGTGCTTCCTTCCTTGTC--------------------------------------------TGG---CCGAT------------------TT-TTCGGTGCC---G-----------------------------------------------CCACCTCTGCCGT-------------CG--CGTCGCGTGCAG-AAAA------------------------------AGTAAGCGCAGAATTAAACGTACACTT-TTAGT------G----------------------------------------------------------------------------G------------------------------------------------------CATTGTTGTT---G-TTT----------------TGGTCCG------TTAGGGTGGCAACCATCGATTTGCGTGTCGCGCAGCCGA--AA--------------------------C-ACAGA--AATTCAAA------ACTCAAAATCAAATCGGCAAAAAGAGTAGACATA--ATAAA---AAGTATATATATCTATATATATATATATAAATA----CAGGGA--AACAAAAGAAATCTAATGCC-------- |
| dm3 |
chrX:6496573-6496950 + |
|
GGCGTTTAGTGA---T---------------TTC----A-----ACCTGTGT------GT-----------GGTGCTTT--------------------GTG----AA-TTTTC-CAAGG-----------------------------------------------------------AATATTG-----GTGTATT--T-TGGCATCGCTGTCGTTTTTTTTTTTG-----------------------------TAGCACACTTACCGTTAT-AATAATAACGTGCTTCCTTCCTTGTC--------------------------------------------TGG---CCGAT------------------TT-TTCGGTGCC---G-----------------------------------------------CCACCTCTGCCACC------------AT--CGTCGCGTGCAG-AAAA------------------------------AGTAAGCGCAGAATTTAACGTACACTT-TTAGT------G----------------------------------------------------------------------------G------------------------------------------------------CATTGTTGTT---G-TTT----------------TGGTCCG------TTAGGGTGGCAACCATCGATTTGCGTGTCGCGCAGCCGA--AA--------------------------C-ACAGA--AATTCAAA------ACTCAAAATCAAATCGGCAAAAAGAGTAGACATA--ATAAAAAAAAGTATATAT--------------------ATA----CAGGGA--AACAAAAGAAATCTAATGCC-------- |
| droEre2 |
scaffold_4690:15427398-15427777 + |
|
GGTGTTTAGTGA---T---------------TTC----C-----AGCTGTGT------GC----------------TCGGCGT---------GCTTCAAGTGAATGAA-TATTAAGAAGGGTA-------------------------------------A------------------AAGATTG-----GTGT--T--T-TTGCATCGCTGTTGTTTT-----TTG-----------------------------TAGCACACTTACCGCTAT-AATAATAACGA----GCTTCCTTGTC--------------------------------------------TGG---CCGATACCGA-------------TT-TTCGGTACC---G-----------------------------------------------C---CTCTGCCGT-----------------CATCGCGTGCAG-AAAA------------------------------AGTAAGCGCAGAATTTAACGTACATTT-TTAGT------G----------------------------------------------------------------------------G------------------------------------------------------CATTGTTGTT---G-TTT----------------TGGTCCG------TTAGGGTGGCAACCATCGATTTGCGTGTCGCGCAGCCGA--AA--------------------------C-ACAGA--AATTCAAA------ACTCAAAATCAAATCGGCAAAAAGAGTAGACATACAATA-----------------CTATATACA------TATATA----CAGGGA--AACAAAAGAAATCTAATGCC-------- |
| droYak3 |
X:2463398-2463757 + |
|
GGTGTTTGGTGA---T---------------TTC----C-----AGCTGTGT------GT----------------------------------TTCAAGTG----AA-TATTAAGAAGG-GA-------------------------------------A------------------AACATTG-----GTGT--T--T-TTGCATCGCTGTTGTTTA-------G---------CACACTGTGTAGC-----TATAGCACACTTACCGCTAT-AATAATAACGT----GCTTCCTCGTC--------------------------------------------TGG---CCGAT------------------TT-TTCGGTACC---G-----------------------------------------------C---CTCTGCCGT-----------------CGTCGCGTGCAG-AAAAAGTAA-----------------------AAAGTAAGCGCAGAATTTAACGTACATTT--AAGT------G----------------------------------------------------------------------------G------------------------------------------------------CATTGTTGTT---G-TTT----------------TGGTCTG------TTAGGGTGGCAACCATCGATTTGCGTGTCGCGCAGCCGA--AA--------------------------C-ACAAA--AATTCAAA------ACTCAAAATCAAATCGGCAAAAAGAGTAGACA-------------------------------------------TA----CAGGGA--AACAAAAGAAATCTAATGCC-------- |
| droEug1 |
scf7180000409522:166889-167246 + |
|
GA-GTG--------------------------------CATATGTGTGGTGT------GTGTGT-------GGTG-----TGTTTTTGGTGTGTTTGCAGTG----AA-TAT---GA------AAAT----------AAA--------------------A---------AAA--A---AA------------------------CATTGCTGTTATTTT-----TTG-----------------------------TAGCACACTTACCGTTAT-AATAATAACGT----GCTTCCTTGTC--------------------------------------------TGG---CCGAT------------------TT-TTCGGTGCC---G-----------------------------------------------C---CTCTCCCGT-----------------CGTCGCGTGCAG-AAAA------------------------------CGTAAGCGCAGAATTTAACGT----TT-TTAGT------G------------CTTTTT----TCGTTGGTTTTTTTGTTT----------------------------------AGTTAT---------------------------------------------------TATTGTTGTTGTTG-TTTTT-------------GTGATCCG------TTAGGGTGGCAACCATTGATTTGCGTGTCGCGCAGTTAA--AA--------------------------C-ACAGA--AATTCAAA------AATCAAAATCAAA--------------------------------------------------------------------CAGGGAAAAACAAAGGAAATCAAATGCC-------- |
| droBia1 |
scf7180000301760:2907913-2908187 - |
|
TGTGTGT--------------------------------------GCTGTGT------GT-----------G---CTGTGTGTGC--------GGCCAAGTG----AG-----------------------------------------------------------------------------------------------------------------------------------------------------T---------AGCGCTG-------CAACGT----GCTTCCTTGTC--------------------------------------------TGG---CCGAT------------------TT--TCGG-GCC---G-----------------------------------------------C---CTCTGCCGC-----------------CGTCGCGTGCAG-GAAAAGTGGCAGAAAGGCGCAGAAAGGCGCAGA---AAGGCGCAGAATTCCACGTAAGTT---CAGA------G----------------------------------------------------------------------------C------------------------------------------------------CATTGTTGCT---G--------------------CGGCCCG------CT-GGGTGGCAACCATCGATTCGCGTGTCGCGCAGCCAA--AA--------------------------C-AGCGA--AGCTCGAA------AACC-AAATCGAA--------------------------------------------------------------------C-GGAA--GACCAGGGAAATCAAATGCC-------- |
| droTak1 |
scf7180000414549:104462-104776 + |
|
G-TATTTAT----------------------------------------TAT------GT-----------GATG--------------------TGAAGTG----AA-TAT-AAGA------GAAAA--------------------------------AAATAGTAACCAA------AACATTG-----------------------GCTGTTATTTT-----TCG-----------------------------TAGCACACTTACCGCTAT-A---ATAACGT----GCTTCCTTGTC--------------------------------------------TGGATTCCCAT------------------TT-TTT--------------------------------------------------------------TCTCTT---------------TC--CATCGCGTGCAG-AAAA------------------------------AGTAAGCGCAGAATTTAGCGTAAATTT-TTAGT------G----------------------------------------------------------------------------T------------------------------------------------------TATTTTTGTTATTG-TTT----------------TGGCCCG------TTAGGGTGGCAACCATCGATTTGCGTGTCGCGGAGTTAA--AAAAAAAACAA--------AAAAAATAAC-ACACA--AATTCAAA------AATCAAAATC------------------------------------------------------------------------GGGGA--AACAAAAGAAATCAAATGCC-------- |
| droEle1 |
scf7180000491044:381633-381954 + |
|
AGTGTTT-GTGA--------------------------------------GT------GT----------------TTTGTGT-------------GAATTG----AA-TTTTGCGA------CAAT----------AA---------------------A------------------TACATTA-----CTTT-TT--G-TTGCCTAGTTGTTATTTT--TTATTG-----------------------------TAGCACACTTTTTGCTAT-A---ATAACGT----GCTTCCTTGTC--------------------------------------------TGG---CCGAT------------------TTTTTCGGTACC---G-----------------------------------------------C---CTCTGCCTT-----------------CGTCGCGTGCAG-AAAA------------------------------AGTAAGTGCAGAATTTAACGTACATTT-TCAGCTATTCAG----------------------------------------------------------------------------T------------------------------------------------------TATTGTTGTT---G-TTT----------------TGGTCCG------TGAGGGTGGCAACCATCGATTTGCGTGTCGCGCGGCCAA--AA--------------------------C-ACAGA--AATTCAAA------AATCAAAATCACATC--------------------------------------------------------------------GGGA--AACAAAAGAAATCAAATGCC-------- |
| droRho1 |
scf7180000776942:63780-64102 + |
|
GGTGTGT-G----------------------------------------TGT------G-----------------------TGA--------ATTCAAGTG----AA-TTTTGCGA------CAAT----------AA---------------------A------------------AACATTG-----TTT-ATT--T-TCGCATAGCTGTTGGTTT--T--TTG-----------------------------TAGCACACTTAGCGCTAT-A---ATAACGT----GCTTCCTTGTC--------------------------------------------TGG---CCGAT------------------TT-TTCGGTACC---G-----------------------------------------------C---CTCTGCCTT-----------------CGTCGCGTGCA----------------------------------------------GAATTTACCGTAAGTTT-TTGGC------G----------------------------------------------------------------------------T------------------------------------------------------TATTGTTGCT---G-TTT----------------TGGTCCG------TTAGGGTGGCAACCATCGATTTGCGTGTCGCGCAGCCAA--AA--------------------------C-ACAGA--AATCCAAA------AATCCAAATCAAATCGGCAAAGAAAACACAAAAA--ATAAA---------------------------------A----AACGGGGA--AACAAAAGAAATCAAATGCC-------- |
| droFic1 |
scf7180000454072:915372-915726 - |
|
TGTGTG--------------------------------C-----TGCTGTGT------GTGT----------GTGTTGTGTGT---------GTTTTAAGTG----AT-GTCCGCGA------CAAT----------AA---------------------A------------------AACATTG-----GTT--TT--T-TGGCATCGCCGTCGTTTT-----TCG-----------------------------TAGCACACTTACCGCTAT-AAT-ATAACGT----GCTTCCTTGTC--------------------------------------------TGG---CCGAT------------------TT-TTCGGTACC---G-----------------------------------------------C---CTATGCCGT-----------------CGTCGCGTGCAG-AAAA------------------------------AGTAAGCGCAGAATTTAGCGTAGTTTT-TTAGT------G----------------------------------------------------------------------------T------------------------------------------------------TATTGTTGCT---G-TTTTT--------------TGGTCCG------TTAGGGTGGCAACCATCGATTTGCGTGTCGCGCAGACAAAAAA--------------------------C-ACAGA--AATTTAAA------AATCAAAATCAAATCGGCAAACAGAG-AGAAA-A--ACAA----------------------------------A----AACGGGGA--AACAAAAGAAATCAAATGCC-------- |
| droKik1 |
scf7180000302367:29711-30122 - |
|
ATTGTGCTG-----------------------------------TGCTGTGT------GTGTGT----------G---TGTGTGTGT-GTGTGTGTGTAGTG----CG----TGAGAGAG-TGCAAA---------------------------------AAACGGTTACCG---------CCTTG-----------------------G------A-------------------------------------------TATTGGTACCGTTATTGGTAATAACGAACGTGCTTCCTTGTC--------------------------------------------TGGGC-TCGAAAAAAAAATTCGCTGCTGTTT-TTC------------------------------------------------------------------------------------G--GGTCGCGTGCAG-AAAA----------------------------AAAGTAAGCGCAGAATTTAACGTAAATTT--CAGA------GTGTTTTTTTGTGTAGTTTTTCTCCTTTTTTGTT---------------------------------------------------------------------------------------------------TTTGTTATT---G-CTTACTGACTGACTGTTT-TC-TCCGGTTTAAGGAGGGTGGCAACCATCGATTTGCGTGTCGCGCTGCCGAA-AA--------------------------C-ACTGAAAAATCCCAA------AATCAAAATCAAATCATCAACACAA-TGGCGAAA-------AAAAAA----------------------TATAAATA----AAAATA--AACAAAAGAAATCAAATGCC-------- |
| droAna3 |
scaffold_12929:3066376-3066760 + |
|
GAAGTGT-GTGTATATTTCTTAAATATTTTTTTT----C-----GGCTGATT------GT------TTTGTGGTTTTCT--------------------GTG----AA-CTTTGCGA------CAAAAGCTGAAAAGCAA--------------------C---------AAA--A---AG-------------------C-TGGCACCGTTGGCGTTTT-------T-----------------------------TAGCACATTTACCGCTAT-A-TAATAACGT----GCTTCCTTGTC--------------------------------------------TGG---CCGAAA-----------------TT-TGCAATTT-----------------------------------------------TTTTTCCACTTTTTTTTA------A------AGCAAGTCGCGTGCAG-A-AA------------------------------AGTAAGCGCAGAATTTAACGTAAGTTT---ATT------G-----TTTATTGTAGTTTTTTTTTTTA------------------------------------------------------------------------------------------------------------------------TT----------------TTTTTTA------TTAGGGTGGCAACCATCGATTTGCGTGTCGCGCAGAAAA--AA--AAAACCAAAAACCAACAACAATAGTAACCGA--AACTC------------CGAAATCGAATC--------------------------------------------------------------------GGGA--AACAAAAGAAATCAAATGCC-------- |
| droBip1 |
scf7180000396036:671-980 - |
|
G-TGAG--------------------------------------TGAAGTGT------AAGATTGTTTTGTGGTTTTCT--------------------GTG----AACTTTTGCGA------CAACA---AAACAACAA--------------------C---------AAAA-A---AG-------------------C-TGGCACCGTTGGCGTTTT-------T-----------------------------TAGCACATTTACCGCTAT-A-TAATAACGT----GCTTCCTTGTC--------------------------------------------TGG---CCGAAA-----------------TT-TTCATTTT----T-----------------------------------------------T---TTTTTTC----------------GCAAGTCGCGTGCA----------------------------------------------GAATTTAACGTAAGTTT---ATT------G----------------------------------------------------------------------------T------------------------------------------------------T---TCTTTT---T-TTT----------------TTTTTTA------TTAGGGTGGCAACCATCGATTTGCGTGTCGCTCAGAAAA--AACAACAACAA--------CAACAAT-----------AATCCAAACTTTAACTCCAAA-----AT-------------------------------------------------------------------CGGGGA--AACAAAAGAAA-------CC-------- |
| dp5 |
XL_group1a:297656-298117 + |
|
TATTTTT-----------CTGCCATGTGTTTT-TTTTC------------TT------GT-----------GCTG--------------------TGGTGTG----AA-TG----GG------CAAT----------AACAGACTCTGACTGCGCTGCAAC---------TACTCCTCCAG-----CTCAAGTTTTCTAC-----A------GTT---TT---TTTTG---------CAC-------------------ATACATATACCGTTTT-A---ATAACGTGCT-GGTTCCCTGTC--------------------------------------------TGGTGTCCGAA------------------TT--ACGGAGCCACTGCCGACGCTGTCAGTGTCGCACTCGCTGTTTCTACGGC-------TGC-CACCGCAGCCACCTCTGTCGCTGTTCG--GGTCGCGTGCAG-A-AA------------------------------AGTAAGCGCAGAATAAACCATACAAATATTAGT------G------------CAGTTTTTTTTCTTCTCTGTATGTGTGTGTGTGTGTGTG----------T-GTGTGTGC-ATGGTG---------------------------------------------------------------TGTGC-G----------------T-----------ATGGAGGGTGGCAACCATCGATTTGCGTGTCGCTCAGCCAA--AA--------------------------C-ACAGCCGCATTCAAA------AATCTAAATCGAATCA---AAG-------------------------------------------------------CTAACGGGAA--CACAAAAGAAATCAAATGCC-------- |
| droPer2 |
scaffold_26:370335-370798 - |
|
TGC-------------------CATGTGTTTT-TTTTC------------TT------GT-----------GCTG--------------------TGGTGTG----AA-TG----GG------CAAT----------AACAGACTCTGACTGCGCTGCAAC---------TACTCCTCCAG-----CTCAAGTTTTCTAC-----A------GTT---TT---TTTTG---------CAC-------------------ATACATATACCGTTTT-A---ATAACGTGCT-GGTTCCCTGTC--------------------------------------------TGGTGTCCGAA------------------TT--ACGGAGCCACTGCCGACGCTGTCAGTGTCGCACTCGCTGTTTCTACGGC-------TGC-CACCGCAGCCACCTCTGTCGCTGCTCG--GGTCGCGTGCAG-A-AA------------------------------AGTAAGCGCAGAATAAACCATACAAATATTAGT------G------------CAGTTTTTTTTCTTCTCTGTATGTGTGTGTGTGTGTGTGTGTGTGTGTGT-GTGTGTGC-ATGGTG---------------------------------------------------------------TGTGC-G----------------T-----------ATGGAGGGTGGCAACCATCGATTTGCGTGTCGCTCAGCCAA--AA--------------------------C-ACAGCCGCATTCAAA------AATCTAAATCGAATCA---AAG-------------------------------------------------------CTAACGGGAA--CACAAAAGAAATCAAATGCC-------- |
| droVir3 |
scaffold_12928:5792784-5793073 + |
|
G----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAGCACATTTACCGTTAT-A---ATAACGT----GCTTCCTTGTCGTTGAACGTCTGTTTTCGTTTGTTTGTGTGCGTTTTCATCTGTTTGC---GCGAT------------------TT-TTCGG-----------------------------------------------------------------------------CGTTTG--CGTCGCGTGCAG-CAAA------------------------------AGTAAGCGCAGAATTTAACGTAATTTT---CGT-----------------------------------------------------------------------------CGATAGTTTTTGCAGCTGTTGTTGTTGTCGCTGCTGCTGCTGCTGCTGTTGTTGTTGTTGTTGTTGTTGTTGTTG-TT----------------G-----T------TGTAGGGTGGCAACCATCGATTTGCGTGTCGTGCAGTCAA--A---------------------------C---------------------------AAATCAAA--------------------------------------------------------------------------------------------TGGAAAACGCGA |
| droMoj3 |
scaffold_6359:4301223-4301400 + |
|
-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGAT------------------TT-TTCGT-----------------------------------------------------------------------------CGTCTG--CGTCGCGTGCAA-C-AA------------------------------AGTAAGCGCAGAATTTAACGTAATTCC--------------------------------TTTTCTTTTTTTTTTTTATATATGTATATATT----------TTTTATATTCGATAGTT---------------------------------------------------------------GTTGC-T----------------G-----------TTATAGGGTGGCAACCATCGATTTGCGTGTCGCGCAGTCAA--A---------------------------C---------------------------AAATCAAA--------------------------------------------------------------------------------------------TGGAAAACGCGA |
| droGri2 |
scaffold_14853:4925569-4925966 + |
|
GTTGTTTCGTGT---T---------------TTC----T-----TCCTATATATATATAT-----------ATTTATTT----------------------------------------------------------------------------TTTAAT---------TTTTCTTCGAG-----TGTGTGTGTTTTTTGTTGCTGTTGTTGTTGTTTT--T--TCGTCACTTTAGCGAAAC--GTATCCAAAATTTAGCACATTTACCGTTAT-A---ATAACGT----GCTTCCTTGTCGATAGAC---------------------------------------G---TCGG-------------------CT-TTTGTTATT---GTTGCTGCTGTTATTGTTGTTCATTTTTCGATTTTGGCAATTTTTTTT-TGTTGTATCT-TTTTTTTTGGCGTCTG--CGTTGCGTGCAGCAAAA------------------------------AGTTAGTGCAGAATTCAACGTAAGTTT---AGT------T----------------------------------------------------------------------CGATAGTTATTGC------------------------------------------------TGTTGTCGTTGTTG-TT----------------G-----T------TGCAGGGTGGCAACCATCGATTTGCGTGTCGCGCAGTCAA--A--------------------------------------------------------------------------------------------------------------------------------------------------CAAATCAAATGCTAAACGCGA |