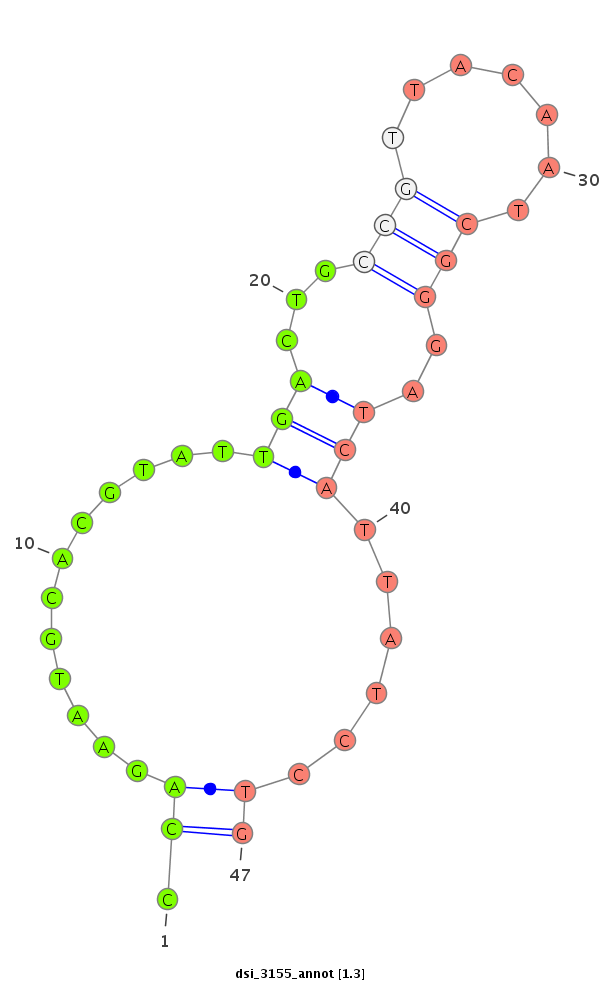
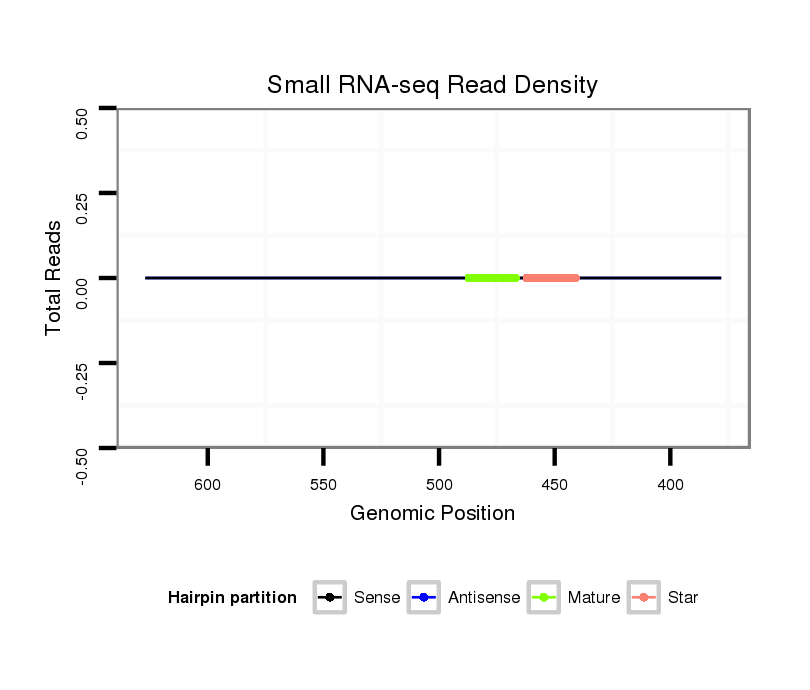
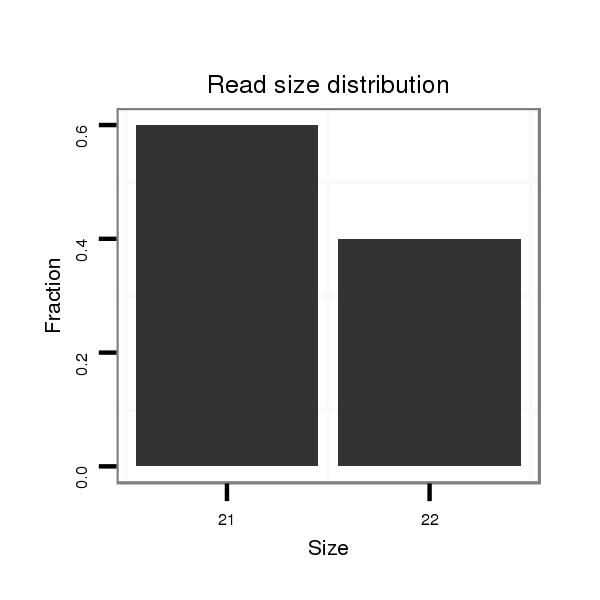
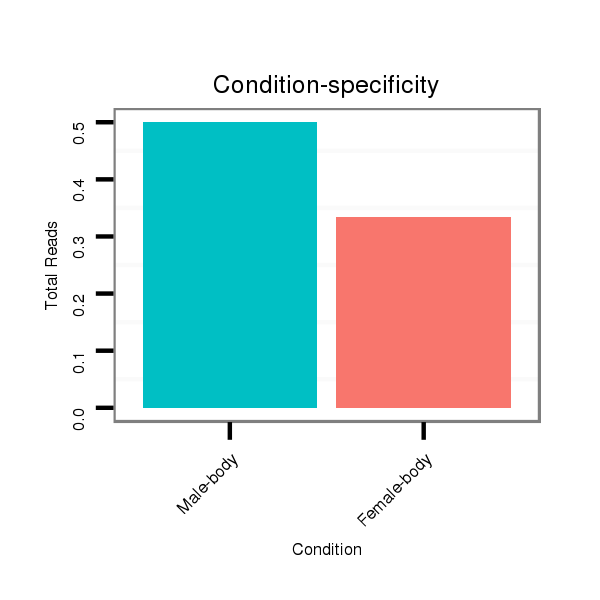
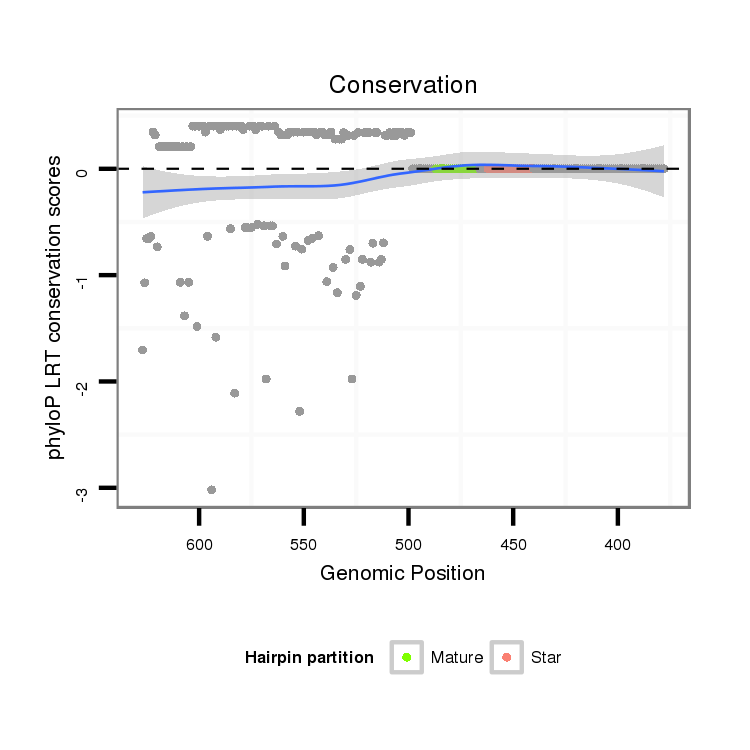
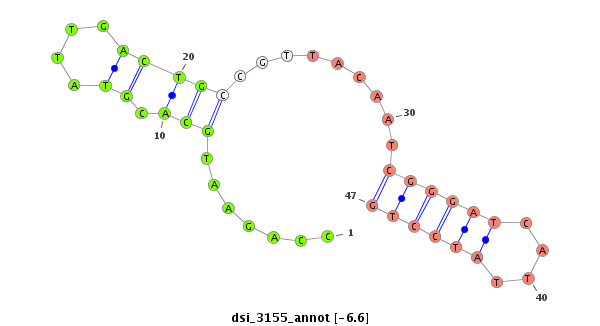
ID:dsi_3155 |
Coordinate:node_18131:428-577 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -6.6 | -5.8 | -5.8 | -5.8 |
 |
 |
 |
 |
intergenic
No Repeatable elements found
| mature | star |
|
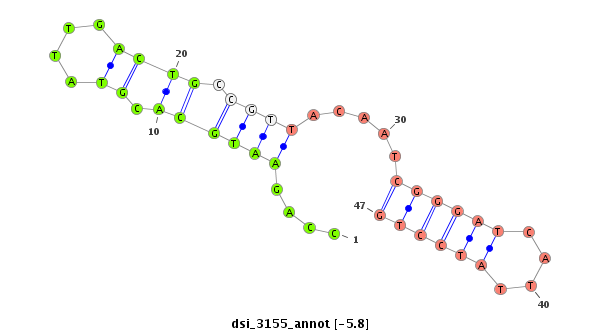
TCCGTTGCATATATTTATATAAAATTTATATATTTCACAAATCTGTTTGCCAGAGAATTCATTTTAGTCCATTAATTTTTTCTCATTGTCACAGATAAGCTGCAAAAATAAATTTCGCTCAGAATCATTTGGTTTTTCGCCCAGAATGCACGTATTGACTGCCGTTACAATCGGGATCATTATCCTGCTGCTCCTTGAACATATTTCGGAAAACTTCGACATTGGAAAACTGAACACCAATAAAGAAAAG
********************************************************************************************************************************************.((............(((...(((.......)))..)))......))*************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
M053 female body |
SRR553488 RT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
M023 head |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................CCAGAATGCACGTATTGACTG......................................................................................... | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................TACAATCGGGATCATTATCCTG............................................................... | 22 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................GACCTTTGAAAACTGAAC............... | 18 | 2 | 8 | 0.25 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................TGACTGCCGGTACAA................................................................................ | 15 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................TCATTTGGTCTGTCTCCCAG.......................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................TATTTAGGAGAACTTCGTCA............................. | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................ATGTCGCTCAGAATCATGAG....................................................................................................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................................................TTCTGCTTCTTGATCATATTT............................................ | 21 | 3 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................TACTTCGACAGTGGAAAA..................... | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................AAATATTTCGGAAAACT................................... | 17 | 1 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................................GCTCAGAATCACTGGG...................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
|
AGGCAACGTATATAAATATATTTTAAATATATAAAGTGTTTAGACAAACGGTCTCTTAAGTAAAATCAGGTAATTAAAAAAGAGTAACAGTGTCTATTCGACGTTTTTATTTAAAGCGAGTCTTAGTAAACCAAAAAGCGGGTCTTACGTGCATAACTGACGGCAATGTTAGCCCTAGTAATAGGACGACGAGGAACTTGTATAAAGCCTTTTGAAGCTGTAACCTTTTGACTTGTGGTTATTTCTTTTC
***************************************************************.((............(((...(((.......)))..)))......))******************************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902009 testis |
SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
M024 male body |
M025 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................GTGCATAACTGCCGGCGAAG.................................................................................. | 20 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................AAAGCGAATCTTAGTCAAGCAA.................................................................................................................... | 22 | 3 | 5 | 0.40 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| .........................................................AAGGTAAATCAGGTAATTAA............................................................................................................................................................................. | 20 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................CCAGAAAGCGGGGCTGACG..................................................................................................... | 19 | 3 | 19 | 0.21 | 4 | 0 | 0 | 0 | 2 | 1 | 1 |
| ..................................................................................................................................................................GGAATGTTAGCCCTGGT....................................................................... | 17 | 2 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................GTGTCTACTCGGTGTTTTTAT............................................................................................................................................ | 21 | 3 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................TAAAAGCACGTCGAGGAACT.................................................... | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | node_18131:378-627 - | dsi_3155 | T------------------------------------------------------------------------------------------------------------------------------------------------CCGTTGCATATATTT-----ATATA---AAATTTA---TATATTTCACA-AATCTGTTTGCCAGAGAATTCATTTTAG-TCCATTAATTTTTTCTCATTGTCACAGATAAGCTGCAAAA-ATAAATTTCGCTCAGAATCATTTGGTTTTTCGCCCAGAATGCACGTATTGACTGCCGTTACAATCGGGATCATTATCCTGCTGCTCCTTGAACATATTTCGGAAAACTTCGACATTGGAAAACTGAACACCAATAAAGAAAAG |
| droSec2 | scaffold_13:108252-108400 - | A-----------------------------------------------------------------------------------TGAGT-TGGGGAAGATGATTCAGGGATTTAATCTATTTTAAAACAGGAAACG--CTCTTACCCGTTGAATATATTT-----ATATT---AAATTTA---TATATATTACA-AATCTGTTTGCCAGAGAATTAATTTTAG-TCCATTAAGCTTCTCTCATTGTCA---------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| dm3 | chr3R:21322728-21322871 - | A--------------------------------------------------------------------------------------------------TGATTCAGGGATCTAATCATTTTTAAAACAGGAAACG--CTCCTATTCGTTGAATATATTTCATTTATTTACAGATATTTA---TATATTTCACA-AATCTCTTTGCCAGAGAATTCATTTAAGGTCCATTAAGTTTCTCTCATTGGCA---------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEre2 | scaffold_4820:6723454-6723653 + | GTTA---TTATAAAAATTAATTTTTTTGTTTAAAACTCTTTCAAGTTCTCTTTATTATAA-------AAATTGCATCACAACAATGAGT-TGGACCAGATTGTTCAGA-GTCAAAGAAATTGCAAAACAGGAAATG--A-------------------------------------TTAATATTA--TATTACAAAATTTGTTTGCCAGAGAATAAAAAT---------------------------------AGGCCGCACGA-ATTTATACTGCTCAGAATCATT------------------------------------------------------------------------------------------------------------------------- | |
| droYak3 | 3R:25161253-25161475 + | GTGCAGGTTAAAAAAATA-------------------------------------TATAGAATTTCGCAATGGCATCATAGCAATGAGTTTGAAGCTGATGATTCAGA-ATCAAATCAATTTTAAAACAGGAAACGAGCATCCATTTAATGA------------------------TTCAAATTATTTCTTACA-AATTTTTTTGTTAAAGTATTAATTTTAG-AGCATTCATTTTCTTTCGTTGGCATATATAAGTAGGAAAATATATATTTTGCTCAGAATCATT------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 05/19/2015 at 03:24 AM