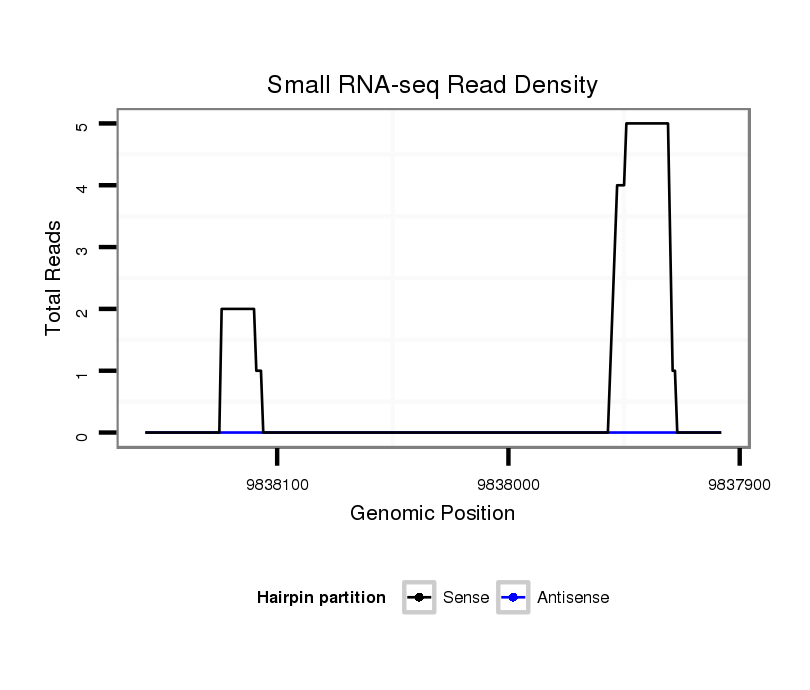
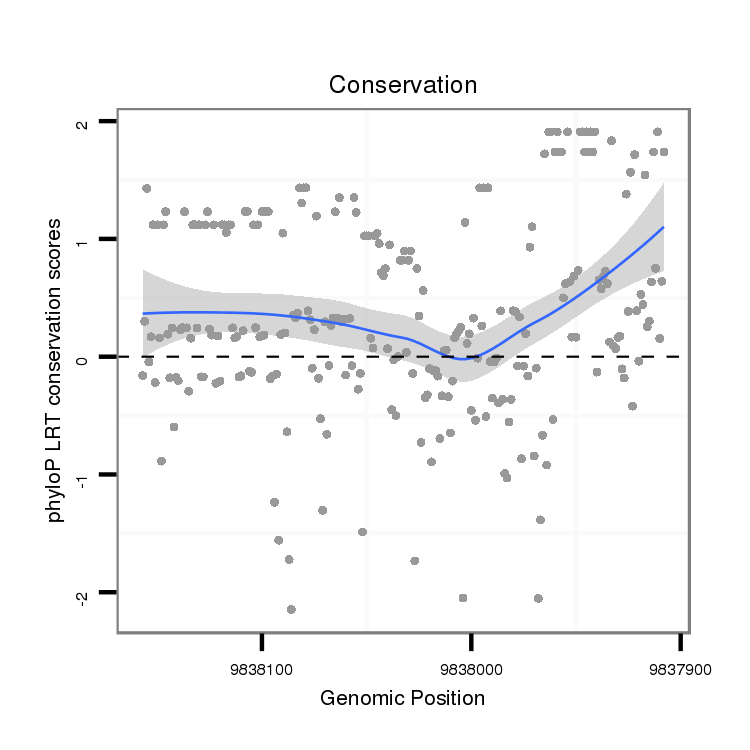
ID:dsi_30867 |
Coordinate:x:9837958-9838107 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
intergenic
No Repeatable elements found
|
AGATCCACCTCTGTATGTGTTAGTGGGGCGGATCGGTTGGGGGTGGCTAAATAGAGGAGTTCCCGAACAGACAAAACAATAACCAGCGTGCTGAAAACAATTTGTCGGGAGCACCACCCACTTGCAGCTGACTAGCCTTGCCTTGCTACGCCTGCACCTATACTCAATTAAATGCCTATCTACGCTTTTCTATTTTGCAGCCCACCAGAAAGACTGAAACAAGGAGCTTCGGCCCAAAAGCGGCGATCCC
**************************************************((((((((((((((((.((((..........((....))...........))))))))))............(((((.((..((((........)))))).)))))...........................)))))))))).......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
GSM343915 embryo |
SRR553488 RT_0-2 hours eggs |
M024 male body |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................GGCGGATCGGTTGGAGGT.............................................................................................................................................................................................................. | 18 | 1 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CCACCAGAAAGACTGAAACAAGGAGC....................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................CGGTTGGGGGTGGCTAAA....................................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................CGGTTGGGGGTGGCT.......................................................................................................................................................................................................... | 15 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................CCAGAAAGACTGAAACAAGGAGCTTC.................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................ACCAGAAAGACTGAAACAAGGAGCT...................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................AAAGACTGAAACAAGGAGC....................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................AAGCCCACCAGAAAGACTGAAACA............................. | 24 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................CACCAGAAAGACTGAAACAAGGAGCT...................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................TTGTCGGGGGCACCA...................................................................................................................................... | 15 | 1 | 16 | 0.94 | 15 | 0 | 0 | 0 | 0 | 15 | 0 | 0 |
| .................................................................................................CAATTTGTCGGCATCACC....................................................................................................................................... | 18 | 2 | 4 | 0.50 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| .....................................................................................................................................................GCCTTCACCTATTCTCAA................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......................................................GGAGTTCCCGATCGGTCAAA............................................................................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................ATAGAGGAGTGCTCGAACA..................................................................................................................................................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................TTTGTCGGGGGCACCA...................................................................................................................................... | 16 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................CTTGCATCTGACTAG................................................................................................................... | 15 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................CCGGATCGGTTGGGG................................................................................................................................................................................................................ | 15 | 1 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................CTAAAGAGTGGAGTTCC........................................................................................................................................................................................... | 17 | 2 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................ATCTGTCGGGACCACC....................................................................................................................................... | 16 | 2 | 18 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................ACCTTGTCGGGTGCACCA...................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
TCTAGGTGGAGACATACACAATCACCCCGCCTAGCCAACCCCCACCGATTTATCTCCTCAAGGGCTTGTCTGTTTTGTTATTGGTCGCACGACTTTTGTTAAACAGCCCTCGTGGTGGGTGAACGTCGACTGATCGGAACGGAACGATGCGGACGTGGATATGAGTTAATTTACGGATAGATGCGAAAAGATAAAACGTCGGGTGGTCTTTCTGACTTTGTTCCTCGAAGCCGGGTTTTCGCCGCTAGGG
**************************************************((((((((((((((((.((((..........((....))...........))))))))))............(((((.((..((((........)))))).)))))...........................)))))))))).......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
M024 male body |
M025 embryo |
M023 head |
SRR553485 Chicharo_3 day-old ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
GSM343915 embryo |
M053 female body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................CGGGGATATGAGTTAATA............................................................................... | 18 | 2 | 1 | 13.00 | 13 | 0 | 7 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................CGGGGATATGAGTTAA................................................................................. | 16 | 1 | 2 | 9.50 | 19 | 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................CGGGGATATGAGTTAAG................................................................................ | 17 | 2 | 9 | 5.33 | 48 | 48 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................GGGGATATGAGTTAA................................................................................. | 15 | 1 | 5 | 5.20 | 26 | 5 | 0 | 0 | 0 | 12 | 7 | 1 | 1 | 0 | 0 |
| .........................................................................................................................................................CGGGGATATGAGTTA.................................................................................. | 15 | 1 | 6 | 5.00 | 30 | 30 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................GGCGGGGATATGAGTTAA................................................................................. | 18 | 2 | 5 | 1.60 | 8 | 1 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 |
| ...AGGTGCAGACATACACAA..................................................................................................................................................................................................................................... | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................CGGGCGGGGATATGAGTTAA................................................................................. | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................CGGGGATATGAGTTAATAC.............................................................................. | 19 | 3 | 7 | 0.71 | 5 | 0 | 3 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................ACGGGGATATGAGTTA.................................................................................. | 16 | 1 | 3 | 0.67 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................AGGGCTTGTCTGTTAT.............................................................................................................................................................................. | 16 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................CGGGGATATGAGTTAAGT............................................................................... | 18 | 2 | 4 | 0.50 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................CGGGGATATGAGTTAGTTCA............................................................................. | 20 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................ACGGGGATATGAGTTAAG................................................................................ | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................CGATGCGGATGTGGATA......................................................................................... | 17 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................CGTGGATATGAGTTAAGG............................................................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................CCACTGCTCGGAACGG............................................................................................................ | 16 | 2 | 20 | 0.30 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 |
| .............................................................................................................................TCCACTGCTCGGAACGG............................................................................................................ | 17 | 2 | 9 | 0.22 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| .....................................................................................................................................................AAGACGGGGATATGAGTTAA................................................................................. | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................................AGCTCCTCAAGGGCTAGTCA................................................................................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................CGGGGATATGAGTTAACTG.............................................................................. | 19 | 3 | 10 | 0.20 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................CGCCACAAGGGCTTGTCT................................................................................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................GGGCGGGGATATGAGTTAAG................................................................................ | 20 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................GTGGGTGAATCTCGACTAA..................................................................................................................... | 19 | 3 | 18 | 0.11 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................CGGGGATATGAGTTACTAT.............................................................................. | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................GTGGGTGAATCTCGACTTA..................................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................GGGGGGTCTTTCGGTCTTTGT............................. | 21 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................CGGGGATATGAGTTACTGT.............................................................................. | 19 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................GTGGGTGAATCACGACTGAT.................................................................................................................... | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................CGTGGATATGAGTTAAGAC.............................................................................. | 19 | 3 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................AGCGGGGATATGAGTTAAT................................................................................ | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................................................................................................AGGGGGATATGAGTTA.................................................................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | x:9837908-9838157 - | dsi_30867 | AGATCCACCTCTGTATGTG----TTAGTGGGG----CGGATCGGTTGGGGGTGGCTAAATAGAGGAGTTCCCGA----------------ACAGACAAAACAATAACCA-----------G-CGTGCTGAAAACAA--TTTGTCGGGAGCACCACCCACTTGCAGCTGACTAGCCTTGCCTTGCTACGCCTG--CACCTATACTCAA-TTAAATGCCTA----------TCTACGCTTTTCTATTTTGCAGCCCACCAGAAAGACTGAAACAAG---------------------GAGCTT---CGGCCCAAAAGCGGCGATCCC----------------------------------- |
| droSec2 | scaffold_15:1073374-1073627 - | AGATCCACCTCTTTATGTG----TTAGTGGGG----CGGATCGGTTGGGGGTGGCTAAATAGAGGAGTTCTTGA----------------ACAGACAAAACAATAACCA-----------G-CGTGCTGAAAACAA--TTTGTCGGGAGCACCACCCACTTGCAACTGACTAGCCTTGCCTCGCTACGCCTG--CACCTATACTCAA-TTAAATGCCTA-----TCT-ATCTACGCTTATGTATTTTGCAGCCCACCAGAAAGACTGAAACAAG---------------------GAGCTT---CGGCCCAAAAGCGGCGATCCC----------------------------------- | |
| dm3 | chrX:10366130-10366371 - | AGATCCACCTCTGTAAGCG----TTAGTGGTG----CGGATCGGTTGGGGGTGGCTAAATAGAGGAGTTCCCGG----------------ACAC-----ACAATAACCA-----------G-CGTGTTGAAAACAA--TTTGTCGGGAGCACCACCCACTTGCAACTGACTAGCCTCGCCTCGCTACGCCTG--CACCTATACTCAA-TTAAATGCCTA----------TCTACGCTTTTGTATTTTGCAGCCCACCAGAAAGACTGAAACAAG---------------------GAGCTT---CGGCCCAAAAG---CGATCCC----------------------------------- | |
| droEre2 | scaffold_4690:4762333-4762572 + | AGATCCACCTCTGTATGCG----TTAGTGGTG----CGGATCGGTTGGGGGTGGCTAAATAGAGGAGTTCCCGA----------------ACACTAAAAACAAAGGCCA-----------G-TGTGCAGTAAACAA--TTTGTAGGGAGCACCACCCACTTGCAACTGCC----------TCGCCTCGCCTC--CACCTGTACTGAA-ATTAATTGCTA----------TCAACGCTTTTGGTTTTTGCAGCCCACCAGAAAGACTGAAACAAG---------------------GAGCTT---CGGCCGAAAAGCGGCGATCCC----------------------------------- | |
| droYak3 | X:18939694-18939944 - | AGATCCACCTCTGTATGCG----TTAGTGGTG----CGGATCGGTTGGGGGTGGCTAAATAGAGGAGTTCCCGG----------------ACTCGCAAAACAAAGGCCA-----------GTGGTGCAGTAAACAA--TTTGTAGGGAGCACCACCCACATGCAACTGCCTCGCTTCGCCCCGGCTCGCCTT--CACTTGTACTCAA-TTTAATTGCTA----------TCTACGCTTTTGATTTTTGCAGCCCACCAGAAAGACTGAAACAAG---------------------GAGCTT---AGGCCGAAAAGCGGCGATCTC----------------------------------- | |
| droEug1 | scf7180000409538:368412-368670 + | AGATCCGCACCTGGTCACAATAAATAGTGGTG----CGGATCGGTCGGGGGTGGCTAAATAGTGGAGTTCCTAAAGGCAAGCATACCAAAACA--GAAAACAAAAGCCA-----------AAAGTGCAGTAAACAA--ATTCTGGGGAGCAC---TC---------------GACTTATACCGCCTAGCTTG--CACTTGTACTAAA-TTTTAACCGTATAATTTCTTTTATTTGTT--ATTTTGTTGCAGCCCACTAAAAAGACTGATACAAG---------------------AAGCTC---CGGCCCAAGAGCGGCGATCCC----------------------------------- | |
| droBia1 | scf7180000302126:3226191-3226378 - | AGAGCCACCACTGGTCGCA----TTAGTGGTG----TGGATCGCTCGGGGGTGGCTAAATGGAGGAGTTCCCTAACTCAACCGCACCAAAACA--CCAAACAAAACCC---------------GCT-----------------------------------------------------------------------TTGAAATCAA-TTTTATCGCTA----------TC------TTTGACTTTTACAGCCCACTAGAAAGACTGAAACAAG---------------------GAGCTT---CGGCCCACAAGCGGTGATCCC----------------------------------- | |
| droTak1 | scf7180000415394:148259-148446 - | AG--CCACCCCTGGTCGCA----TTACTGGTG----CAGATCGGTCGGGGGTGGTTAAATAGAGGAGTTATTAGAGGCAGGCGCATCC--------AAGACAAAACCCC-----------A-TTCCCAGTAAAAAAAAATTACC-------------------------------------------------------CTAATAAT-CCTAACCGATT-----TCT-ATCTTTT--TCTTCCTCTTGCAGCCCACCAGAAAGACTGAAACAAG-------------------------------------------GCGATCCC----------------------------------- | |
| droEle1 | scf7180000491087:288006-288254 + | AGACCCGCCTCTGATCAAA----TTGTAGGTG----CGGAGCGAGTGGGGGCGTCAAAAAAGAGTAATTCCTAGAGGCAAACGTACTAAAACA--GACAACAAAAGCCT-----------AGTGGGCAGTAAACAA--ATTGTAGGGTGC----CCCGATGGCAACTAC------CCGCCCCACCTTCCTTT--CACTTGCACTCAA-TGTTATCCCCA----------TCTGTG---------CTTGCAGCCCACCAGAAAGACTGAAACAAG---------------------GATCGTCTTTGGCCCAAAAGCGGCGATCCC----------------------------------- | |
| droRho1 | scf7180000777289:23547-23782 - | -GATACGCCTCTGGTCGCA----TTATAGGTG----CGGATTGGGTGG-GGTGGCTAAATGGAGGAGTTCCTAGGGGCAAGCGTTCCACATCA--GACAACAAAAGTCA-----------G-TGGGCAGTAAAGAT--ATTGTAGGGAGCAC---CCAACTGCAACTAGC--------------GCCGTTTC--CTCTTGTACTCAA-TGTTTTCCCTA----------TCTGT---------TCTTTCAGCCCACCAAAAAGACTGAAACAAG---------------------GAGCTT---CAGCCCAAAAGCGGCGATCCC----------------------------------- | |
| droFic1 | scf7180000454050:288858-289073 - | TGATCCGCCCCTGGTTGCA----TTAGAGGTGTATGTAGATCGGGTGGGGGTTGTGCAAAAGAGGAGTTTCCA-AGGCAGGGGTACCAAAACA--TAAAACAAAAG--------------G-CGGGCAGTACTCGA--TT-------------ACCCGATTGCAACTGAC---------------TCGCTTG--CACTTGTACTCAA-TTTTATTGTAA----------ATTCTG---------CTTACAGCTCATCAGAAAGACTGAAACAAG---------------------GAGCTT---C-----------GGCGGTTCC----------------------------------- | |
| droKik1 | scf7180000302698:772602-772800 + | C-----------------------------------------------------------------------------------------------AAAACAAAAGCCACAAAGCCCAATG-TGTGCAGTAAACAA--ATTGCTGGGATCACC----------------------------TGCCCCGCTCGCAC-TCCTTACTCACGCTCCACACCCA----------TCTTCGTT--CCTCTTTTGCAGCTCACCAGGAAGACTGAGGCAAGAA-------------------GATTCC---CAGCCGAAGAGCGGCGATCCCCAACACAAGAAAACGAGAGCGAGATCGAGAGAGAG | |
| droAna3 | scaffold_13335:2346699-2346781 + | TTAT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTATTTTTCAGCTTACCCGAAAGACTGACGACGAATAATTAGCAGCTAGAAGAAGGAGCTT------GCTAAACGCAACGATTCC----------------------------------- | |
| dp5 | XL_group1e:5712891-5712950 - | ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCTGTCTCTTGCAGCTCACCAGAAAGACTGAAGCAAG---------------------AAGATT-----GCCCA--ATCAACGATCCC----------------------------------- | |
| droPer2 | scaffold_21:640626-640685 - | ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCTGTCTCTTGCAGCTCACCAGAAAGACTGAAGCAAG---------------------AAGATT-----GCCCA--ATCAACGATCCC----------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/18/2015 at 10:48 AM