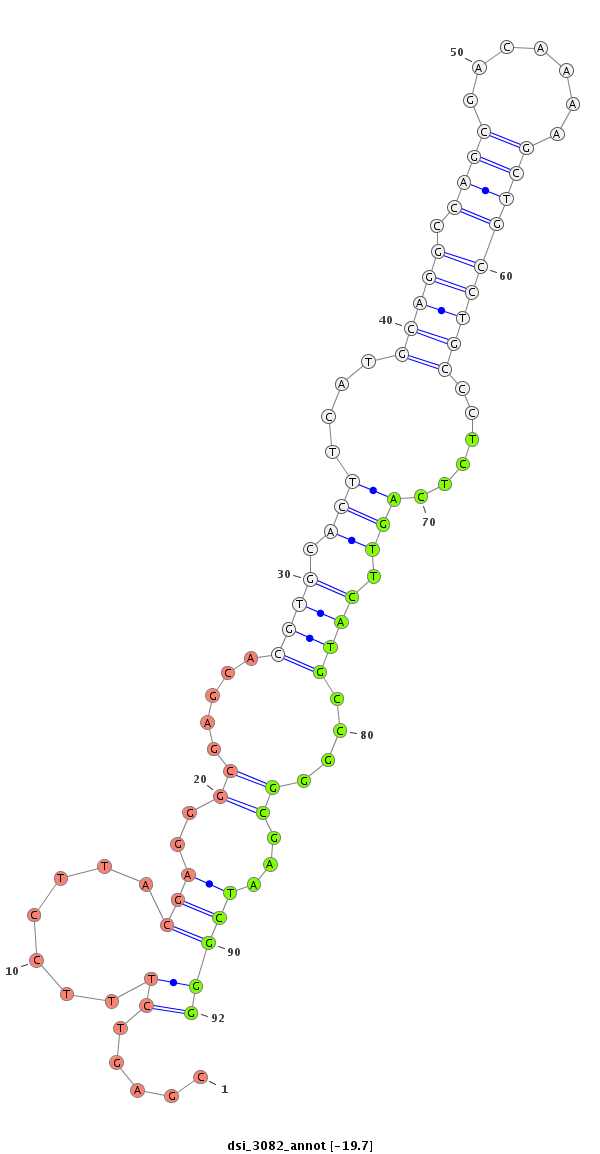
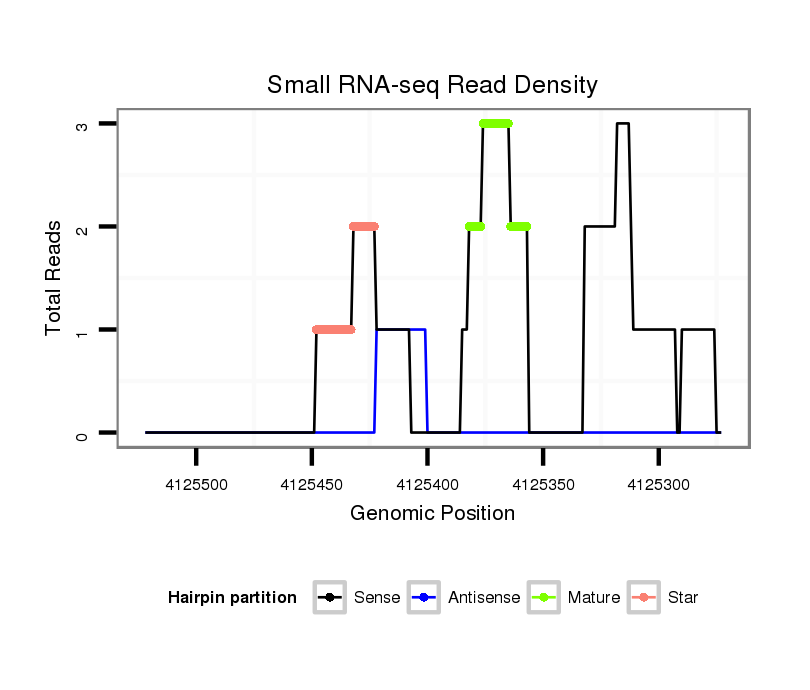
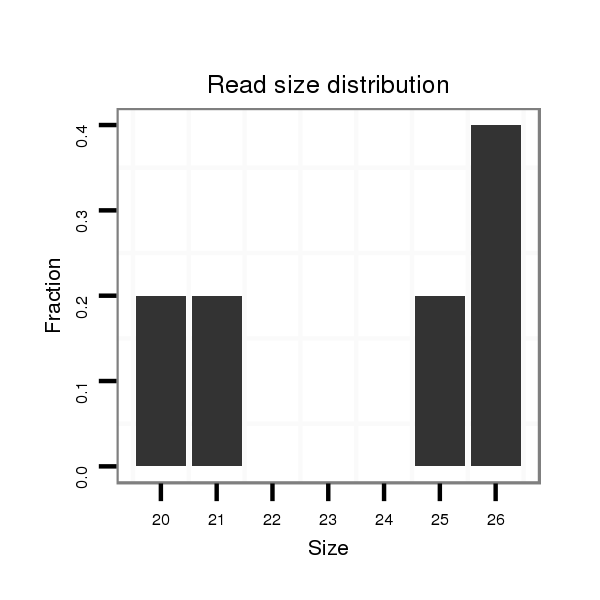
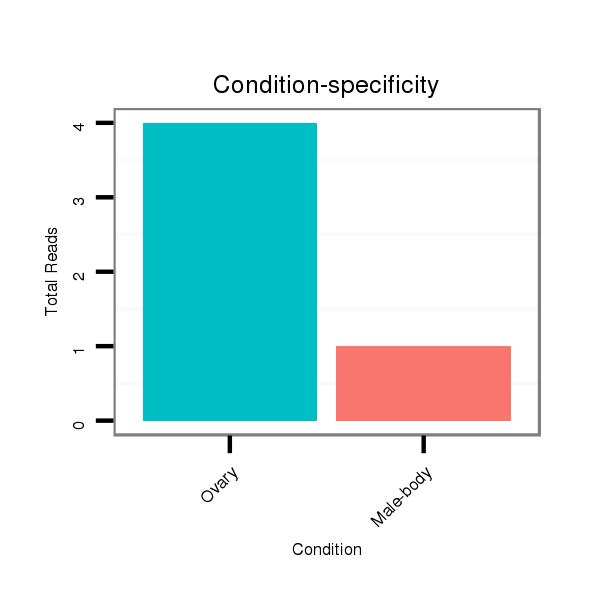
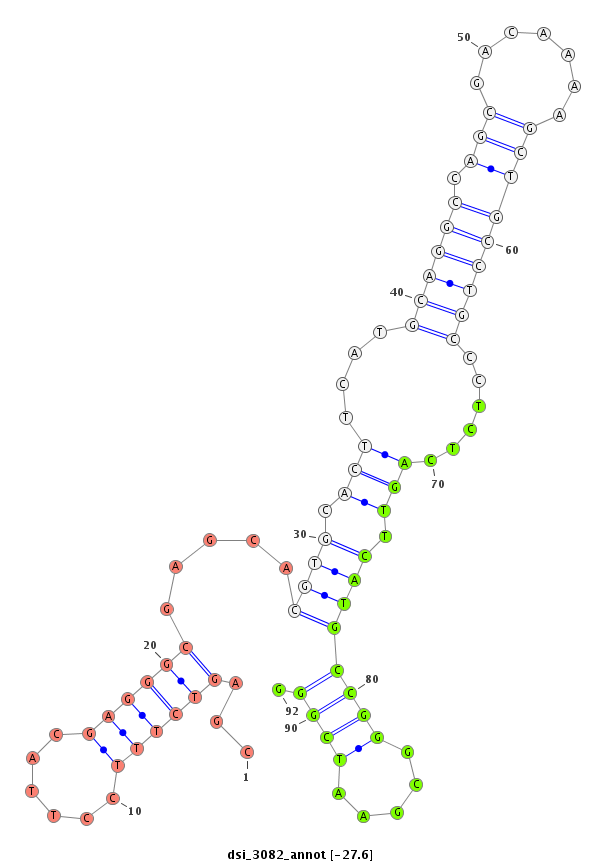
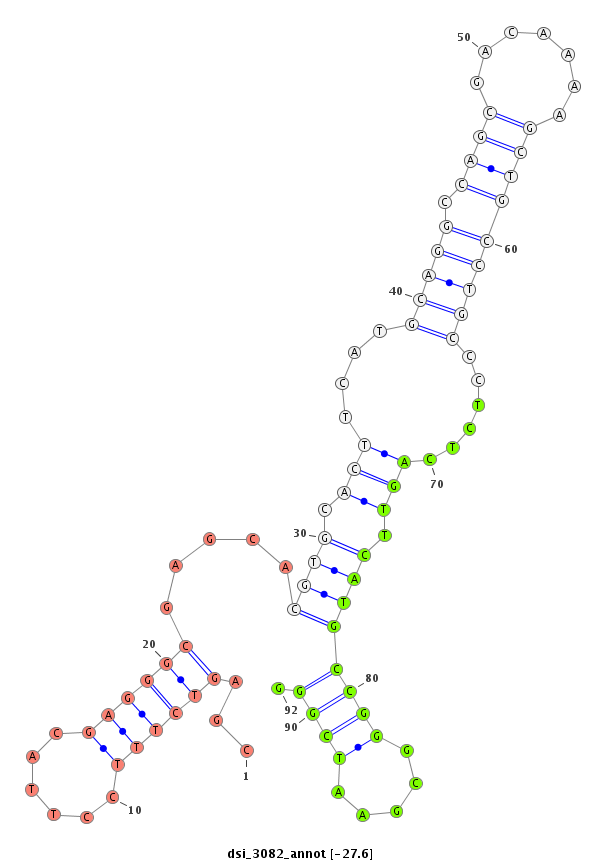
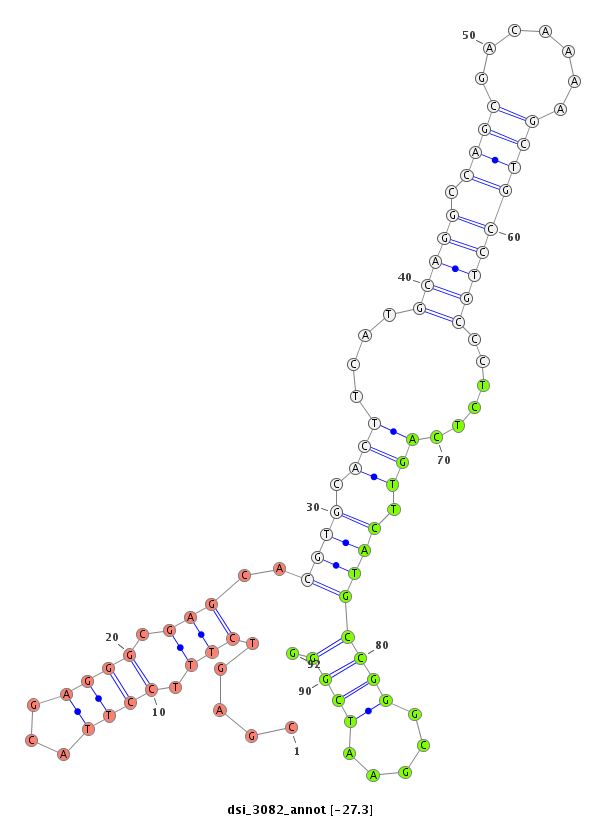
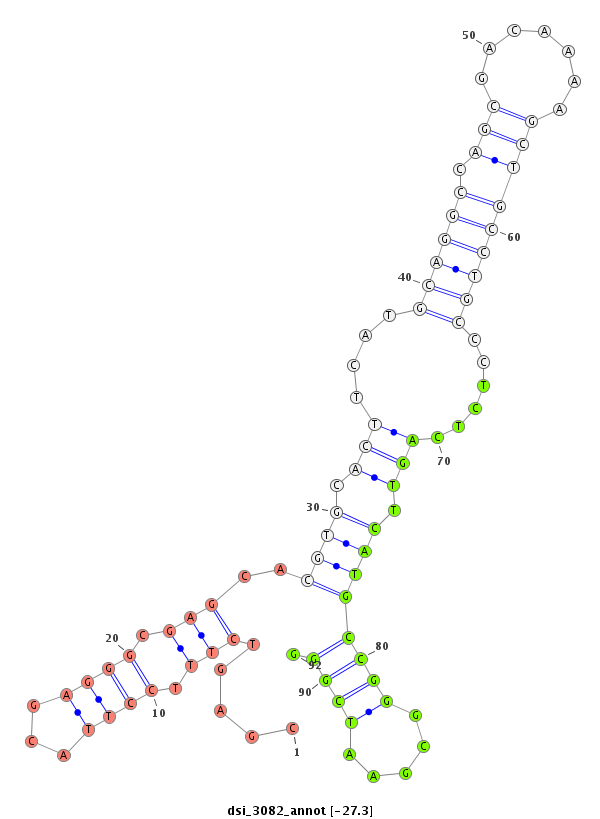
ID:dsi_3082 |
Coordinate:3r:4125323-4125472 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -27.6 | -27.6 | -27.3 | -27.3 |
 |
 |
 |
 |
intergenic
No Repeatable elements found
|
ACTACTCGGTGTTTCAGTGCTGCTACCGTGAACACGGCGAGGAGTGCAAGGTGCGGGTGGTATGCGACCAGAAACGAGTCTTTCCTTACGAGGGCGAGCACGTGCACTTCATGCAGGCCAGCGACAAAAGCTGCCTGCCCTCTCAGTTCATGCCGGGCGAATCGGGAGTCATATCCTCGCTGTCGCCCAGTAAGGAGCTGCTGATCAAGAATACCACCAAGCTCGAGGAAGCCGACGACAAGGAGGAGGA
**************************************************************************.....((.......(((..((.....((((.(((....(((((.((((.......)))))))))......))).))))....))...)))))************************************************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR553487 NRT_0-2 hours eggs |
M024 male body |
M025 embryo |
GSM343915 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ............TTCAGTGCTGCTACCGTGAGCACGGCGA.................................................................................................................................................................................................................. | 28 | 1 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................TAAGGAGCTGCTGATCAAGA........................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................................................................................TCTCAGTTCATGCCGGGCGAATCGGG.................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................CCCTCTCAGTTCATGCCGGGC............................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................TTCATGCCGGGCGAATCGGG.................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................CGAGTCTTTCCTTACGAGGGCGAGCA...................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................CGACGACAAGGAGGA... | 15 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................TGAGGGCGAGCTGGTGCACTTC............................................................................................................................................ | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................TAAGGAGCTGCTGATCAAGAA....................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................AGGGCGAGCACGTGCACTTCATGCA....................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................TCAAGAATACCACCAAGCTCGAGGAA.................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................CCGTGAACTCGGCGA.................................................................................................................................................................................................................. | 15 | 1 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................TGACCAAGGCGAGGAGT............................................................................................................................................................................................................. | 17 | 2 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ACTCCTCGGTGTTGCAG......................................................................................................................................................................................................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
|
TGATGAGCCACAAAGTCACGACGATGGCACTTGTGCCGCTCCTCACGTTCCACGCCCACCATACGCTGGTCTTTGCTCAGAAAGGAATGCTCCCGCTCGTGCACGTGAAGTACGTCCGGTCGCTGTTTTCGACGGACGGGAGAGTCAAGTACGGCCCGCTTAGCCCTCAGTATAGGAGCGACAGCGGGTCATTCCTCGACGACTAGTTCTTATGGTGGTTCGAGCTCCTTCGGCTGCTGTTCCTCCTCCT
************************************************************************************.....((.......(((..((.....((((.(((....(((((.((((.......)))))))))......))).))))....))...)))))************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................CATAGACCTCACTATAGGAGC....................................................................... | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ....................................................................................................GCACGTGAAGTACGTCCGGTCG................................................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ......................................................................................................ACGTGAAGTACGGCCGG................................................................................................................................... | 17 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 |
| ......................................................................................................................................................................................AGGGGGTGATTCCTCGA................................................... | 17 | 2 | 9 | 0.11 | 1 | 0 | 0 | 1 |
| .................ACGATGATGGTACTTGT........................................................................................................................................................................................................................ | 17 | 2 | 14 | 0.07 | 1 | 0 | 1 | 0 |
| .......................................................................................................................................................................................GGGGGTGATTCCTCGA................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 |
Generated: 04/23/2015 at 08:35 PM