
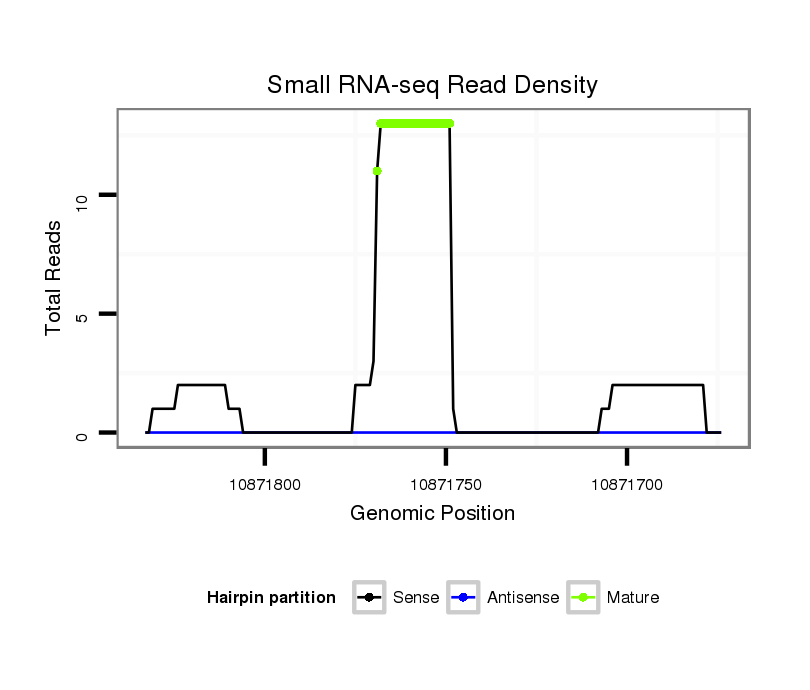
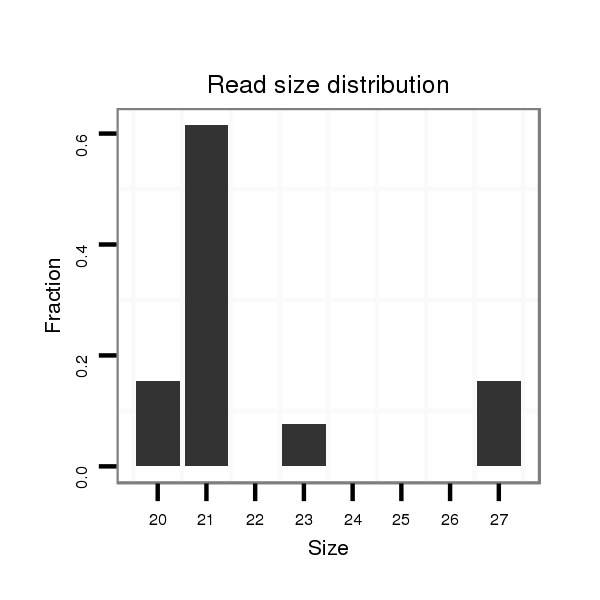

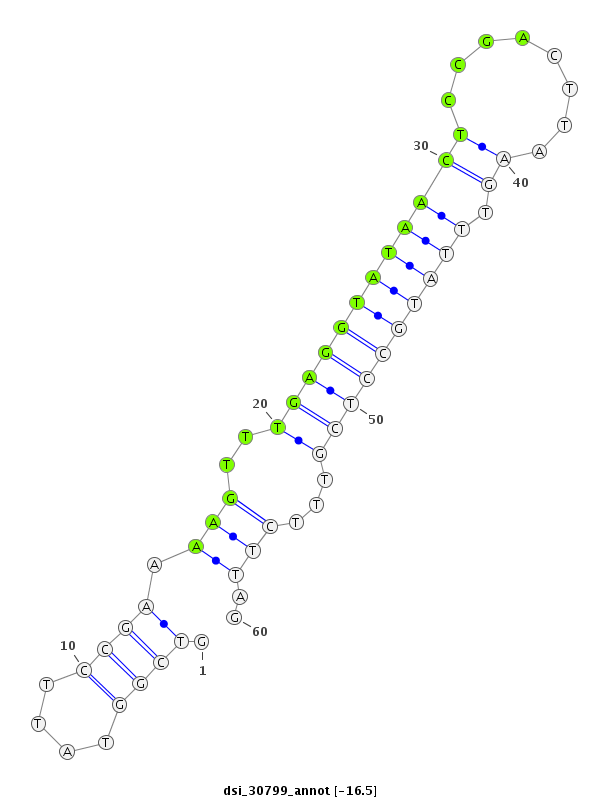
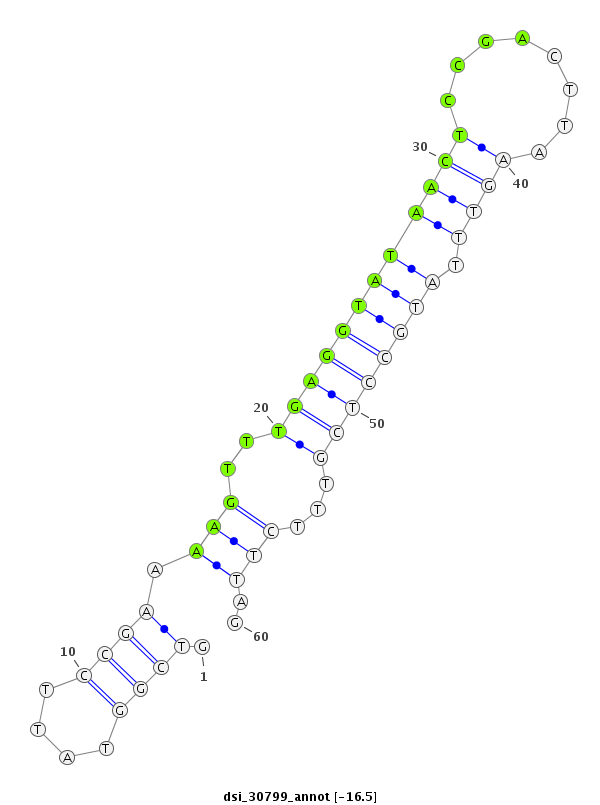
ID:dsi_30799 |
Coordinate:3r:10871724-10871783 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -16.5 | -16.5 | -16.5 | -15.9 |
 |
 |
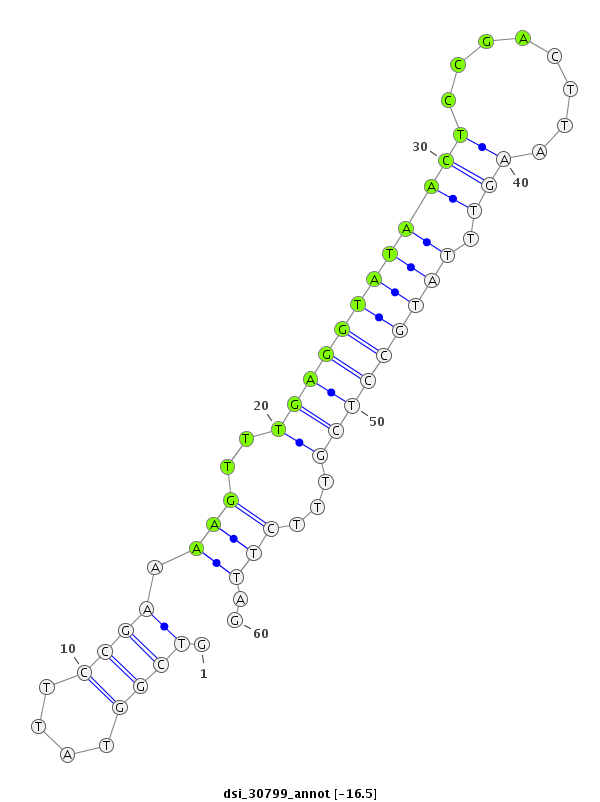 |
 |
CDS [3r_10871553_10871723_-]; exon [3r_10871494_10871723_-]; CDS [3r_10871784_10872068_-]; exon [3r_10871784_10872068_-]; intron [3r_10871724_10871783_-]
No Repeatable elements found
| ##################################################------------------------------------------------------------################################################## TTGCAGCCATTAAGATTCAGGACCCGCCCAATGTACTTCTTACCATGCAGGTCGGTATTCCGAAAAGTTTGAGGTATAACTCCGACTTAAGTTTATGCCTCGTTTCTTAGCAAATGTGTGAGATGCTCATCAGCATTGCCCGGACCTGTGGCATCAAAGT **************************************************.......((..((((....((((((((((((........)))).))))))))))))..))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
M023 head |
SRR553485 Chicharo_3 day-old ovaries |
GSM343915 embryo |
SRR618934 dsim w501 ovaries |
M024 male body |
SRR902009 testis |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................AAGTTTGAGGTATAACTCCGA........................................................................... | 21 | 0 | 1 | 8.00 | 8 | 6 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................AGTTTGAGGTATAACTCCGA........................................................................... | 20 | 0 | 1 | 2.00 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................AGTTTGAGGTATAACTCCGG........................................................................... | 20 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TCCGAAAAGTTTGAGGTATAACTCCGA........................................................................... | 27 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................CTTGAGGTATAACTCCGA........................................................................... | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........TTAAGATTCAGGACCCGC..................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..GCAGCCATTAAGATTCAGGAC......................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................AGCATTGCCCAGACCTGCGGCA....... | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................AAAGTTTGAGGTATAACTCCGAC.......................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................TCAGCATTGCCCGGACCTGTGGCATC..... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........ATTGAGATTCGGGACCCGCCC................................................................................................................................... | 21 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TCCGAAAAGTTTGAGGTATAACTCCGG........................................................................... | 27 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................TCATCAGCATTGCCCGGACCTGTGGCATC..... | 29 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................AAGTTTGAGGTATAACTCCTC........................................................................... | 21 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................TGCCCGGACCTGTCGCACC..... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........GTTAAGATTCCGGAACCGC..................................................................................................................................... | 19 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........GTTAAGATTCCGGACCCT...................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
AACGTCGGTAATTCTAAGTCCTGGGCGGGTTACATGAAGAATGGTACGTCCAGCCATAAGGCTTTTCAAACTCCATATTGAGGCTGAATTCAAATACGGAGCAAAGAATCGTTTACACACTCTACGAGTAGTCGTAACGGGCCTGGACACCGTAGTTTCA
**************************************************.......((..((((....((((((((((((........)))).))))))))))))..))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902009 testis |
SRR618934 dsim w501 ovaries |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|
| ...................TTTGGGCGGGTTACATGAAGA........................................................................................................................ | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .......................TGCGGGTTACATGAAGAATGGT................................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...........................................GTAGGTCCAGCCATA...................................................................................................... | 15 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 |
| ......GGGAATTCTAAGTCCAG......................................................................................................................................... | 17 | 2 | 7 | 0.14 | 1 | 0 | 0 | 1 |
| ...........................................................................................................................................GGCCTGGACTGCGTAG..... | 16 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 |
Generated: 04/24/2015 at 10:51 AM