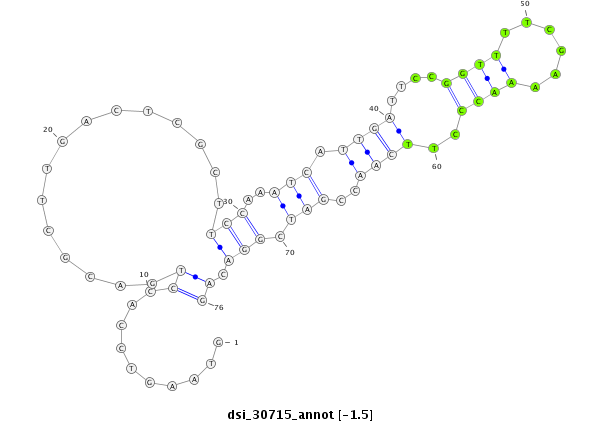
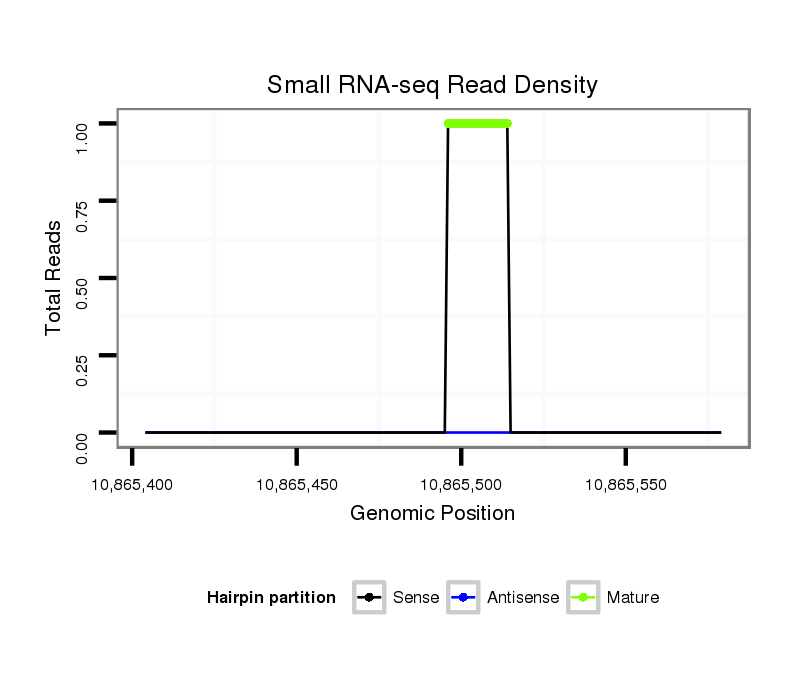
ID:dsi_30715 |
Coordinate:3r:10865454-10865529 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
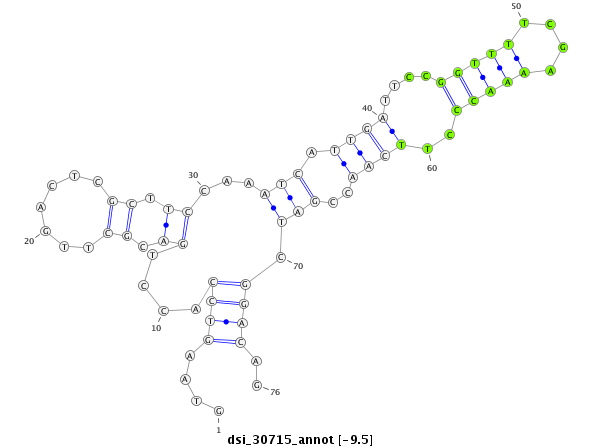
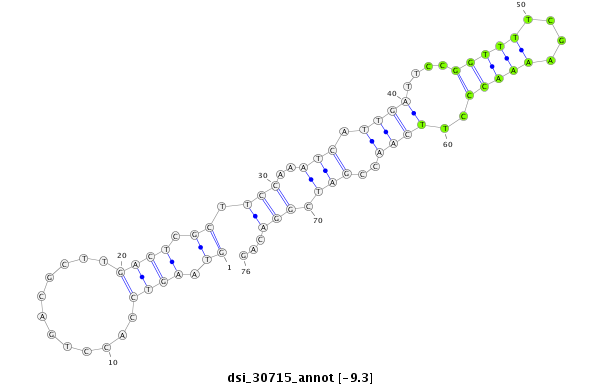
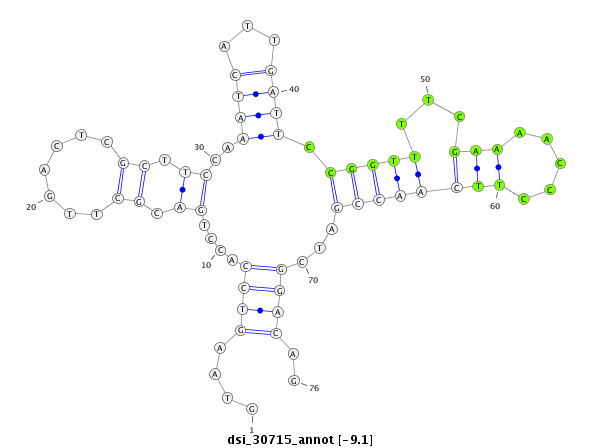
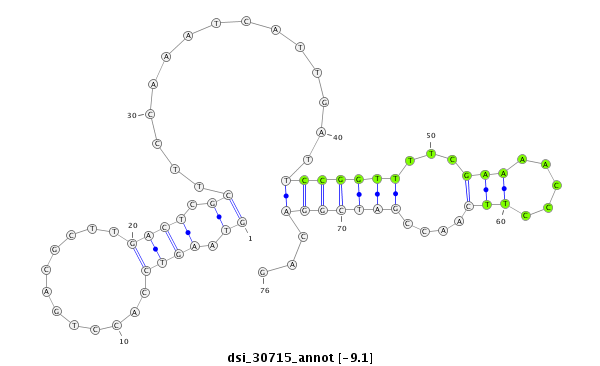
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -9.5 | -9.3 | -9.1 | -9.1 |
 |
 |
 |
 |
exon [3r_10865373_10865453_+]; CDS [3r_10865373_10865453_+]; CDS [3r_10865530_10865817_+]; exon [3r_10865530_10865817_+]; intron [3r_10865454_10865529_+]
No Repeatable elements found
| ##################################################----------------------------------------------------------------------------################################################## ATCAGCAAGCCACCCGCGCCATCAAGTCCGCCGATAGCTTTGAGGAGAAGGTAAGTCCACCTGACGCTTGACTCGCTTCCAAATCATTGATTCCGGTTTTCGAAAACCCTTCAACCGATCGGACAGAAAATGCGCAATCGTTTCACGGTGCTCTTCGAACTCGGTGGAGAATATCA **************************************************..........((...............(((..(((.((((....((((......))))..))))..))).))).))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
M023 head |
O002 Head |
SRR553488 RT_0-2 hours eggs |
GSM343915 embryo |
SRR553487 NRT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................GCCGAAAGCTTTGAGGAG................................................................................................................................. | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................TTCAACCGATCGGACAGT................................................. | 18 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................CCGGTTTTCGAAAACCCTT................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................TGATTCCGGCTTTCGAA........................................................................ | 17 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................GCGCAGATAGCTTTGATGAGA................................................................................................................................ | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................CGATAGCTTTGTGGAGAA............................................................................................................................... | 18 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................GCTCTTCGAACGTGTTGGAGA...... | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................GACAGCTTTGAGGAG................................................................................................................................. | 15 | 1 | 15 | 0.27 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................AGCTTTGACGAGTAGG............................................................................................................................. | 16 | 2 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 |
| ................................GCTAGCTTTGAGGAG................................................................................................................................. | 15 | 1 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CTCCTGAGGCTTGACT....................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................CTCGTGACGCTTGACT....................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......AACCAGCCGCGCCATCA........................................................................................................................................................ | 17 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TAGTCGTTCGGTGGGCGCGGTAGTTCAGGCGGCTATCGAAACTCCTCTTCCATTCAGGTGGACTGCGAACTGAGCGAAGGTTTAGTAACTAAGGCCAAAAGCTTTTGGGAAGTTGGCTAGCCTGTCTTTTACGCGTTAGCAAAGTGCCACGAGAAGCTTGAGCCACCTCTTATAGT
**************************************************..........((...............(((..(((.((((....((((......))))..))))..))).))).))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
M024 male body |
M023 head |
SRR618934 dsim w501 ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
SRR902009 testis |
M025 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................TTTAGCAAAGTGCCAGGACAAG.................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................CCTCTTTGATTCAGGTGGA.................................................................................................................. | 19 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................AATGCGAGCTGTGCGAAGGT............................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................TCAGCAAAGTGCCAGGACAAGC................... | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................GGATGTTGGCTAGCC...................................................... | 15 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........TGGTGGGCGCGGTAGTTAACG................................................................................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................TCAGCAAAGTGCCAGGACAA..................... | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................GCAAAGTGCCAGGACAAG.................... | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................AGCAAAGTGACAGGACAAGC................... | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................................AGTCACCTCTTATAG. | 15 | 1 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................TGAGCGAATGTGTAGTA......................................................................................... | 17 | 2 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................GCAAAGTGCCAGGACAA..................... | 17 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................TTAGCAAAGTGACAGGA........................ | 17 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................TCGAGAAGCTTGAGACA........... | 17 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................CCACGAGAAGCGTGAGA............. | 17 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................ACAAAGTGCCAGGACAAGC................... | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................GGAAGTTGGTGAGCCTGTG.................................................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Generated: 04/24/2015 at 10:50 AM