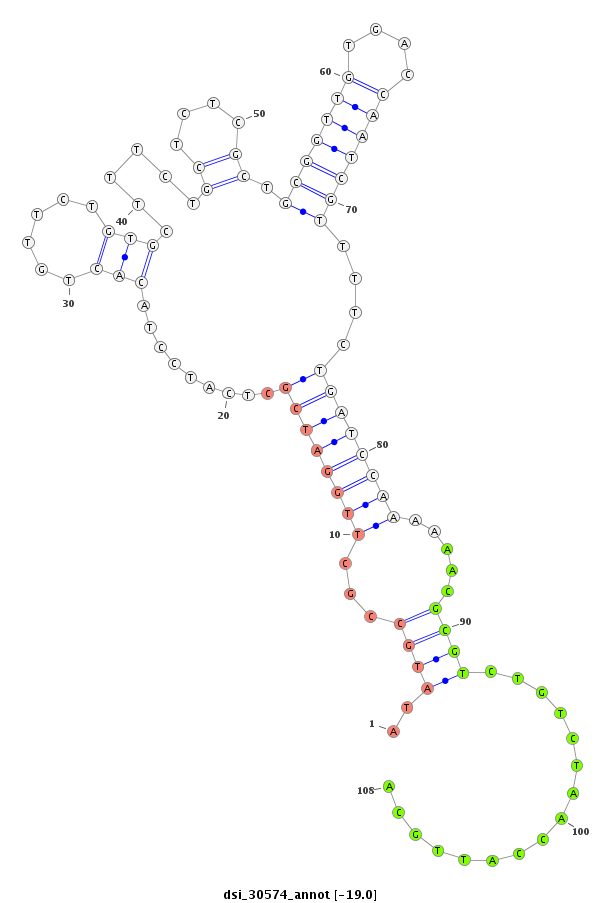
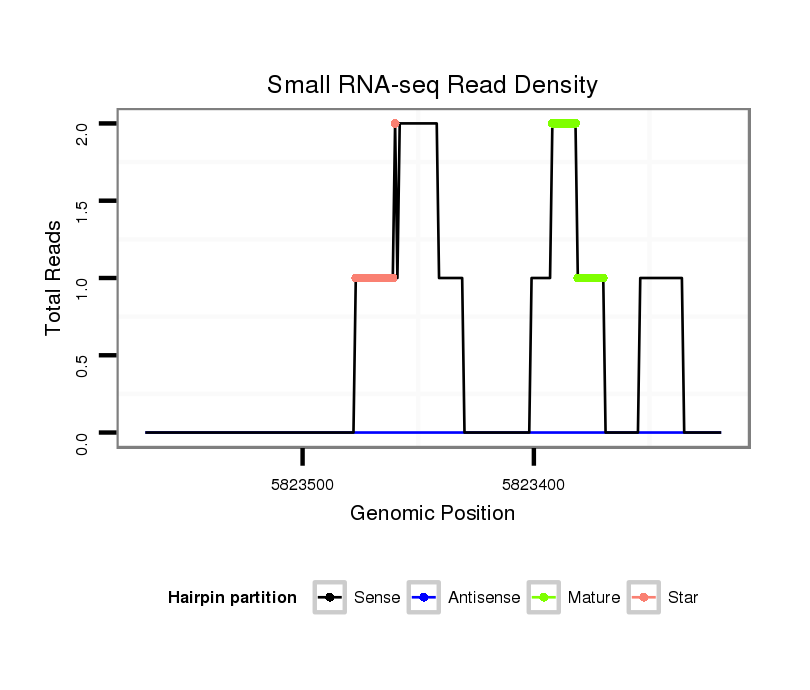
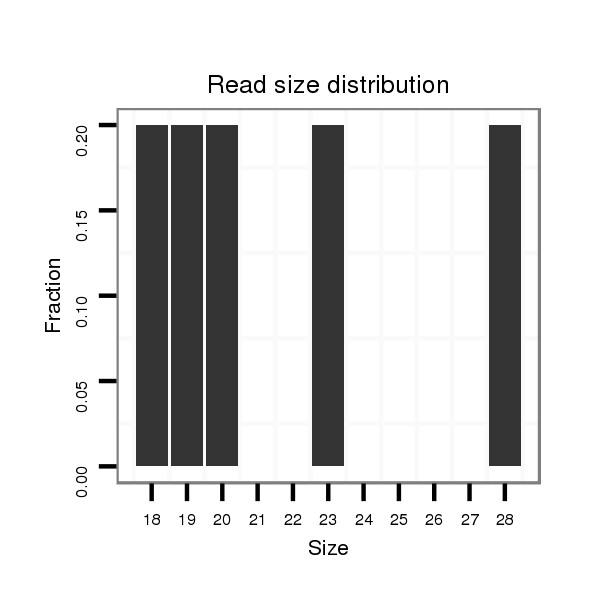
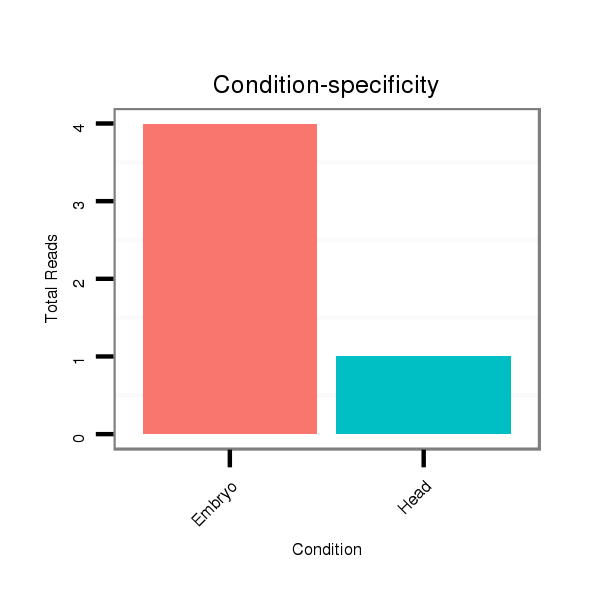
ID:dsi_30574 |
Coordinate:2r:5823369-5823518 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
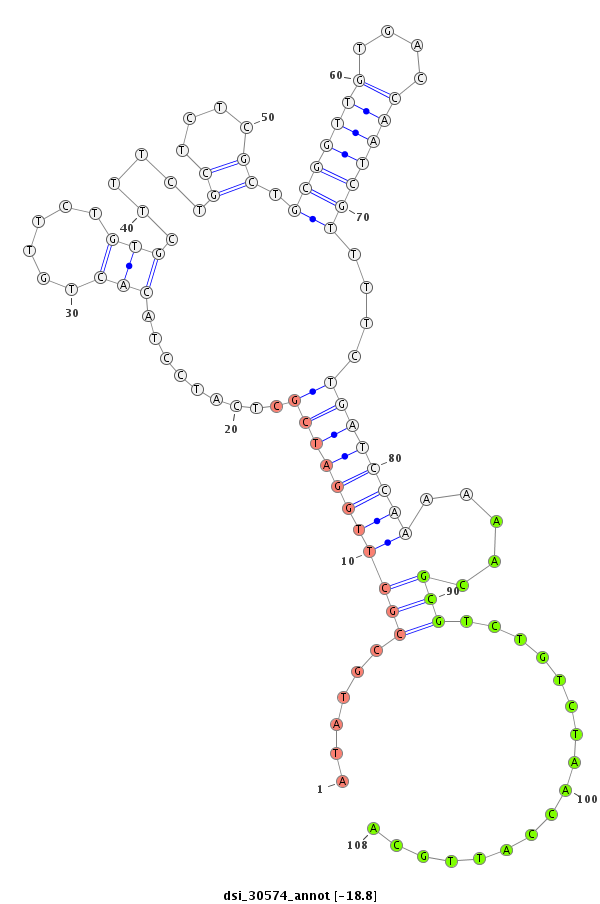
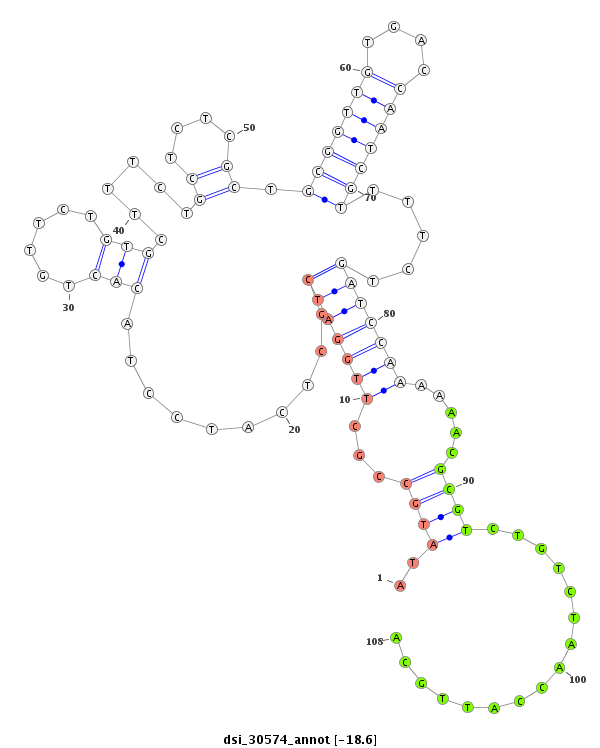
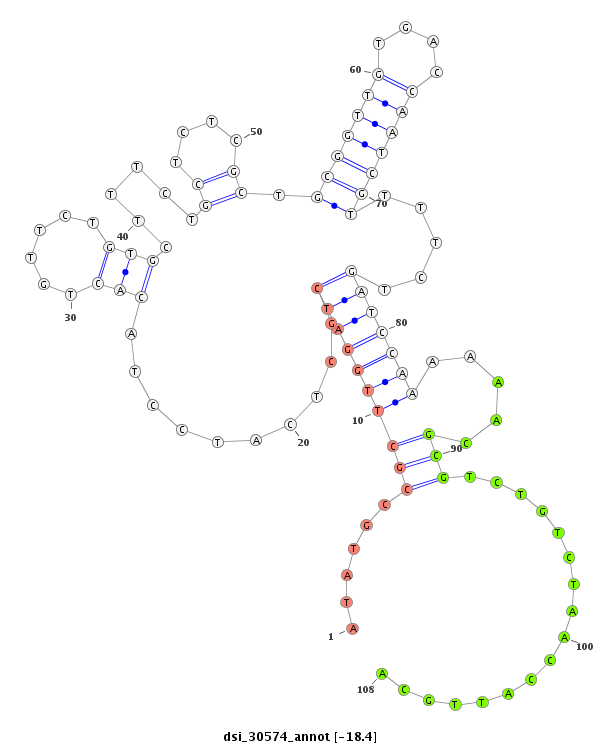
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -18.8 | -18.6 | -18.4 |
 |
 |
 |
intron [2r_5823223_5823335_-]; CDS [2r_5823336_5823368_-]; exon [2r_5823336_5823368_-]; intron [2r_5823369_5824566_-]
No Repeatable elements found
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------#################################----------------- GTGTAGTTAGTCGTAGCTTAAATAACCCATTGGTTGTTCTGCGCATCTCATTCACGCCGTATCTCGCCCGTATCTTGTGTGTCTCTCTTTCATATGCCGCTTGGATCGCTCATCCTACACTGTTCTGTGCTTTCTGCTCTCGCTGCGGTTGTGACCAATCGTTTTCTGATCCAAAAAACGCGTCTGTCTAACCATTGCAGACTAATCCGGCTGTAAAGTCTGCCGTCAAATCGGTGAGTGGTCCAGTCCC *******************************************************************************************..((((...((((((((.........(((......)))......((....)).(((((((....)))))))....)))))))).....))))................*************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
M023 head |
M024 male body |
GSM343915 embryo |
SRR553487 NRT_0-2 hours eggs |
M053 female body |
SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................AACGCGTCTGTCTAACCATTGCA................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................AAAGTCTGCCGTCAAATCG................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................ATATGCCGCTTGGATCGC............................................................................................................................................. | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................GATCCAAAAAACGCGTCTGT............................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................CTCATCCTACACTGTTCTG........................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................CATCCTACACTGTTCTGTGCTTTCTGCT................................................................................................................ | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................GCACTAATCCGGCTGTAAAGTCTGCCGT........................ | 28 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................CTGCCGAGAAATCGGTGAGGG.......... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................ACCAAGCTTTGTCTGATCCAA............................................................................ | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................AAGTCCGCCGTCAAA.................... | 15 | 1 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................ATAACAGATTCGTTGTTCTG................................................................................................................................................................................................................. | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................ATTCGTTGTTCTGCG............................................................................................................................................................................................................... | 15 | 1 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................CTGCGGTTGTCACCA............................................................................................. | 15 | 1 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................CCAGATAACGCGTCTGACT............................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CACATCAATCAGCATCGAATTTATTGGGTAACCAACAAGACGCGTAGAGTAAGTGCGGCATAGAGCGGGCATAGAACACACAGAGAGAAAGTATACGGCGAACCTAGCGAGTAGGATGTGACAAGACACGAAAGACGAGAGCGACGCCAACACTGGTTAGCAAAAGACTAGGTTTTTTGCGCAGACAGATTGGTAACGTCTGATTAGGCCGACATTTCAGACGGCAGTTTAGCCACTCACCAGGTCAGGG
***************************************************..((((...((((((((.........(((......)))......((....)).(((((((....)))))))....)))))))).....))))................******************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
GSM343915 embryo |
M024 male body |
SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................AGAGAGCGGGCATAG................................................................................................................................................................................ | 15 | 1 | 7 | 0.29 | 2 | 2 | 0 | 0 | 0 | 0 |
| .......................................................CGGTATGGAGGGGGCATAG................................................................................................................................................................................ | 19 | 3 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................GATGTAAGAAGAAACGAAAG.................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................AGGTAACGTCTGATT............................................. | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................TCGAGTAGGATGTGGC................................................................................................................................ | 16 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................ACAGAGTGCTGACGTCTGA............................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................................................................................................................................................................CTCAGACGGCAGTGAAGC................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
Generated: 04/24/2015 at 12:37 AM