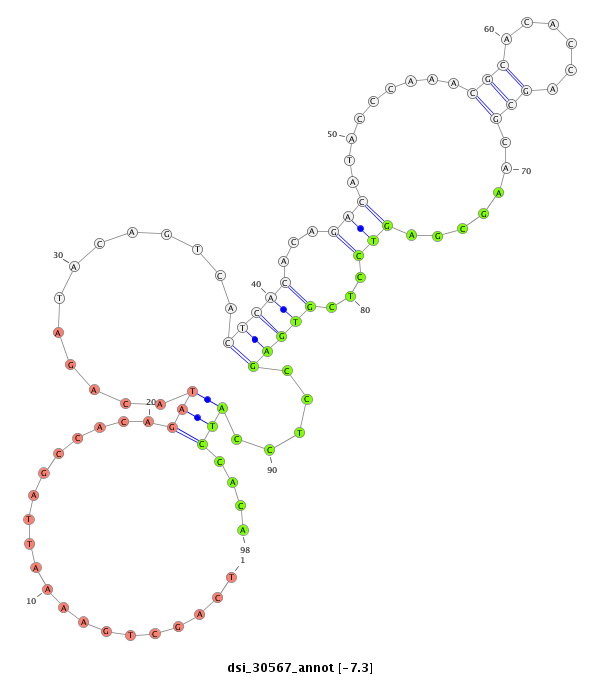
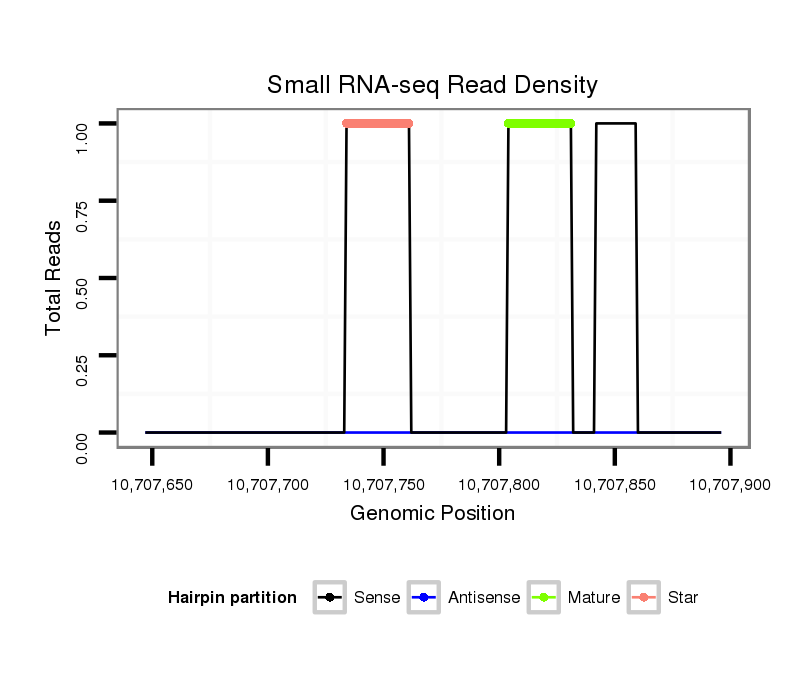
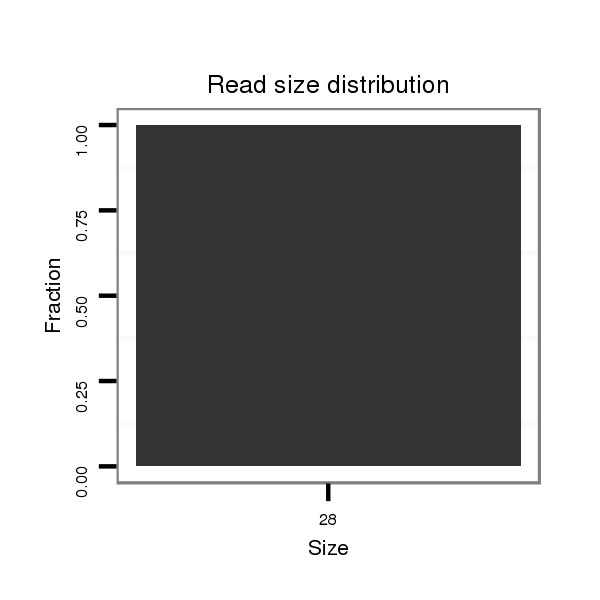
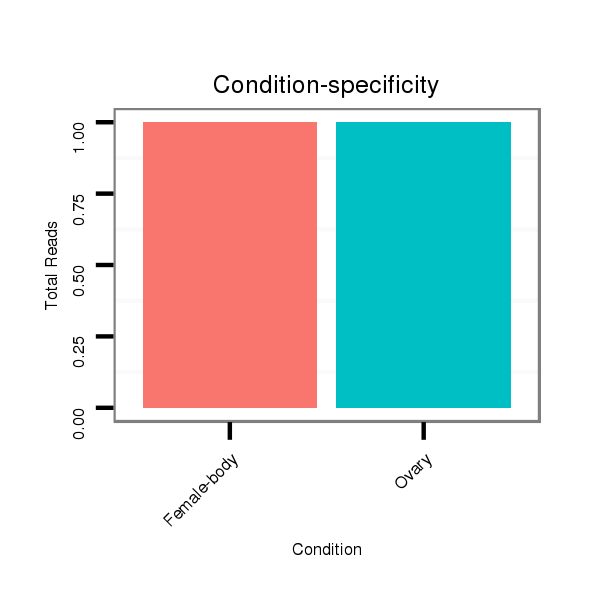
| AG------CAGCATCAACAGCAGCAG------CAGCA---GCAGCA---TCAACA---G---------C-----------------------------------------------AGCAGCAGCAGCAG-CAT--CAACAGCAGCAGCAGCAGCAGCAT------------------C--AA---C---AAC---AGCATCA------------------ACAACAGAAGCAGCA------ACAGCAACTGCAGCTG-------C--AA--------ATGC---------A---GCAACAGCAGC-----------AGCAGAAGAAACTGCAGCAGC---------------------------AACAACA------ACAA------------------------CAACAACAACAAC------------------------------------------AGCAGCAGCAACAACAAC-------------------------------------AACAGC-AGCAGCA--GCA | Size | Hit Count | Total Norm | Total | M046
Female-body | V041
Embryo | V049
Head | V056
Head | M060
Embryo | V110
Male-body |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAACAAC-------------------------------------AACAGC-AGCAGCA--GC. | 22 | 20 | 0.25 | 5 | 0 | 0 | 0 | 0 | 5 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAACAAC-------------------------------------AACAGC-AGCAGC...... | 19 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................CAACAGAAGCAGCA------ACAGCAA.............................................................................................................................................................................................................................................................................................. | 21 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........CAGCATCAACAGCAGCAG------CAGCA---GC............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 25 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........................................CAGCA---TCAACA---G---------C-----------------------------------------------AGCAGCAGCAGC....................................................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................CAG-CAT--CAACAGCAGCAGCAGCAGC............................................................................................................................................................................................................................................................................................................................................................................................ | 25 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................ACAA------------------------CAACAACAACAAC------------------------------------------AGC....................................................................... | 20 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AC-------------------------------------AACAGC-AGCAGCA--GCA | 18 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................CAGCAGCAGCAGCAT------------------C--AA---C---AAC---................................................................................................................................................................................................................................................................................................................................................... | 22 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................GCAG-CAT--CAACAGCAGCAGCAGC............................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................GCAG------CAGCA---GCAGCA---TCAACA---G---------C-----------------------------------------------................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................GCAGCA---TCAACA---G---------C-----------------------------------------------AGCAGCAGC.......................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .G------CAGCATCAACAGCAGCAG------CAGC................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................GCAGCAGCAGCAG-CAT--CAACAGC........................................................................................................................................................................................................................................................................................................................................................................................................ | 23 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................GCAG-CAT--CAACAGCAGCAGCAG................................................................................................................................................................................................................................................................................................................................................................................................ | 22 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................GCAGCA---TCAACA---G---------C-----------------------------------------------AGCAGCAG........................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .G------CAGCATCAACAGCAGCAG------CAG.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........AGCATCAACAGCAGCAG------CAGCA---GCA............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 25 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGCAGCAACAACAAC-------------------------------------AACAGC-AG.......... | 23 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................AGCAG-CAT--CAACAGCAGCA.................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................AACAGCAGCAGCAGCAGC............................................................................................................................................................................................................................................................................................................................................................................................ | 18 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................AGCAGCAGCAGCAGCAGCAT------------------C--AA---C---A........................................................................................................................................................................................................................................................................................................................................................ | 25 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................CAG-CAT--CAACAGCAGCAGCAG................................................................................................................................................................................................................................................................................................................................................................................................ | 21 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| AG------CAGCATCAACAGCAGCA.............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 19 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GCAGCAACAACAAC-------------------------------------AACAGC-AGCAGCA--G.. | 28 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................AGCAGCAG------CAGCA---GCAGCA---TCAACA---............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 25 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................AG-CAT--CAACAGCAGCAGCAGCAGCA........................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............AACAGCAGCAG------CAGCA---GCAGCA---T..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................CAGCA---TCAACA---G---------C-----------------------------------------------AGCAGCAG........................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............ATCAACAGCAGCAG------CAGCA---............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................CAAC------------------------------------------AGCAGCAGCAACAACAAC-------------------------------------AACAGC-A........... | 29 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................................AACA---G---------C-----------------------------------------------AGCAGCAGCAGCAG-CAT--............................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................AG-CAT--CAACAGCAGCAGCAGCA.............................................................................................................................................................................................................................................................................................................................................................................................. | 22 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................A---TCAACA---G---------C-----------------------------------------------AGCAGCAGCA......................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........CAGCATCAACAGCAGCAG------CAG.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................AACAGCAGCAGCAGCAGCAGCAT------------------..................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................AACA---G---------C-----------------------------------------------AGCAGCAGCAGC....................................................................................................................................................................................................................................................................................................................................................................................................................... | 18 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................AA------------------------CAACAACAACAAC------------------------------------------AGCAGCAGC................................................................. | 24 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................CA------ACAA------------------------CAACAACAACAAC------------------------------------------AGCAGCA................................................................... | 26 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........CAGCATCAACAGCAGCAG------C...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............AACAGCAGCAG------CAGCA---GC............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 18 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........AGCATCAACAGCAGCAG------CAGCA---............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................A---G---------C-----------------------------------------------AGCAGCAGCAGCAG-CAT--CAACA.......................................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................AT--CAACAGCAGCAGCAGCA.............................................................................................................................................................................................................................................................................................................................................................................................. | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................AGCA---TCAACA---G---------C-----------------------------------------------AGCAGCAGCA......................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................AGCA---TCAACA---G---------C-----------------------------------------------AGCAGCAGCAGCA...................................................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................A---GCAGCA---TCAACA---G---------C-----------------------------------------------AGCA............................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAGCAACAACAAC-------------------------------------AACAGC-AGCAG....... | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |