ID:dsi_30564 |
Coordinate:3r:10853757-10853906 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
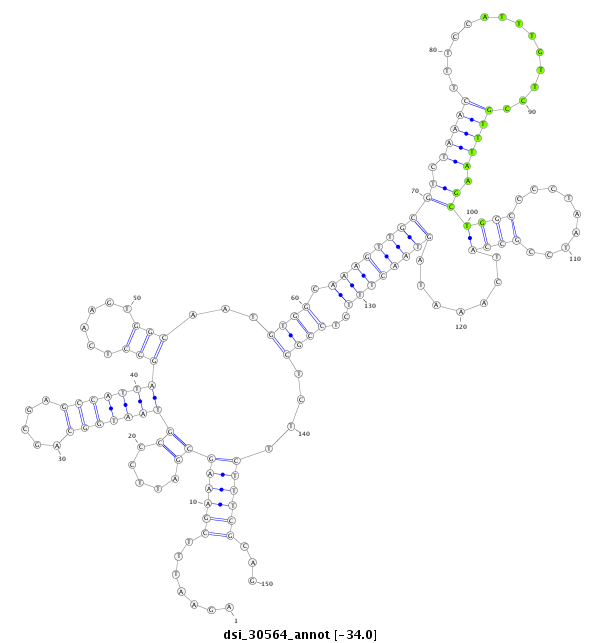
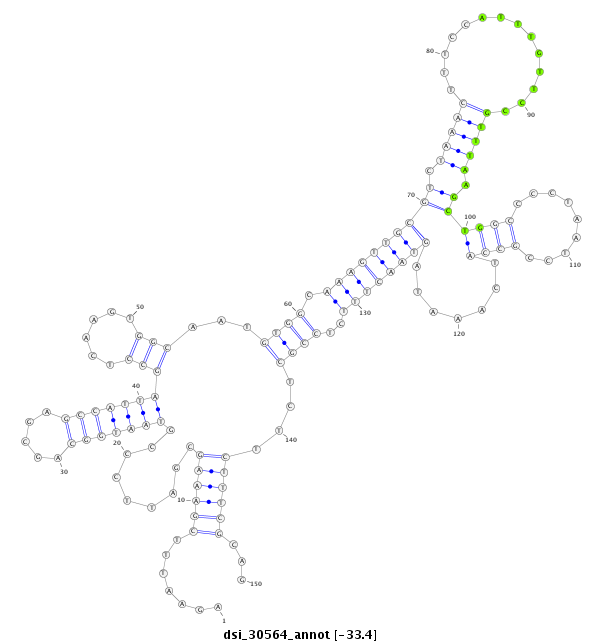
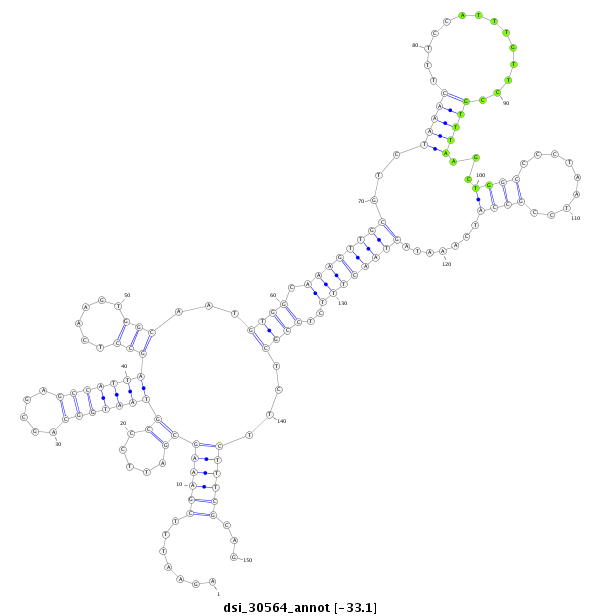
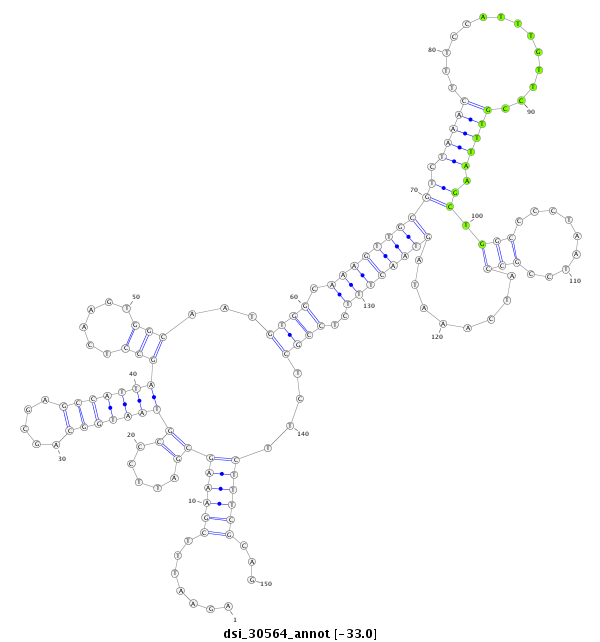
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -33.4 | -33.1 | -33.0 |
 |
 |
 |
intergenic
No Repeatable elements found
|
CGTTTCGCACTCAAAGTTAATGGCTTGGTTAGGCCACGCCCTGCTCATCTAGAATTTCGAAAGCGATTCCCGTAATGGCAGCGAGCCATTAGCCTCAAGTGGCAATGTGGCAAAGTTGCGTCTAAACTTTCCATTTGTTCCGTTTAAGCTGGCCCCTAATCCGCCATCAAATAGTAACTTTCTCCGCTCTTCTTTCGCAGGTTTCTTCAATCAAAGGGAAAAATCAACTGTTAACCGGCGAGTGGACGCA
**************************************************.......((((((((.....))(((((((.....)))))))(((......)))...((((.((((((((((.(((((..............))))).))((((.........)))).......))))))))..))))....))))))...************************************************** |
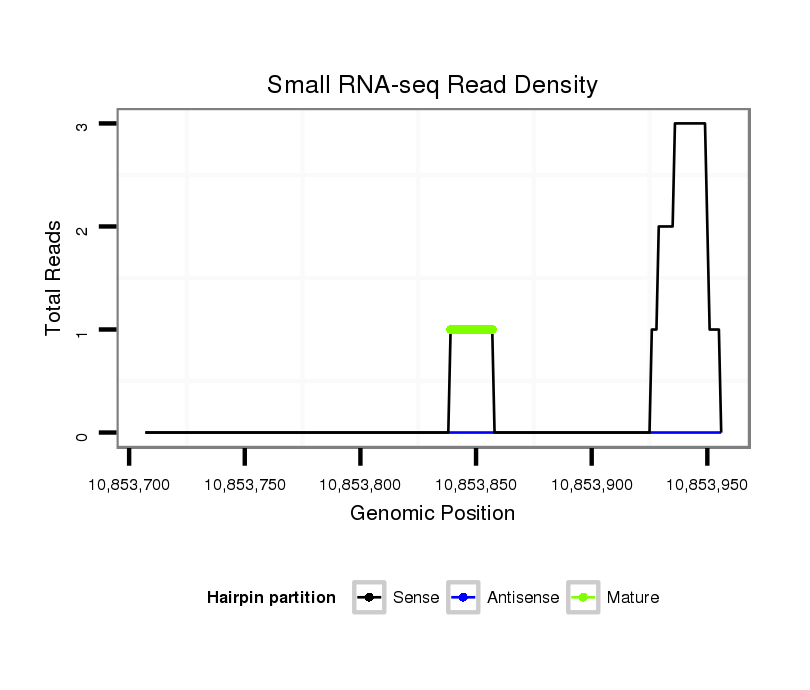
Read size | # Mismatch | Hit Count | Total Norm | Total | M053 female body |
M023 head |
SRR553486 Makindu_3 day-old ovaries |
GSM343915 embryo |
SRR618934 dsim w501 ovaries |
M024 male body |
SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................AGCGATTCCCGTAGTAGCGGCGA...................................................................................................................................................................... | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................AAAATCAACTGTTAACCGGCGAGT....... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................GTTAACCGGCGAGTGGACGC. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................ATCAACTGTTAACCGGCGAGTG...... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................ATTTGTTCCGTTTAAGCTG................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................AATGGCTTAGTGAGGCCTC..................................................................................................................................................................................................................... | 19 | 3 | 11 | 0.91 | 10 | 0 | 2 | 0 | 0 | 3 | 5 | 0 | 0 |
| ...............................................................CGATTCCCGTAGTAGCGGC........................................................................................................................................................................ | 19 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................................................TTTTAACCGTCGCGTGGAC... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................TAAGCTGGCCCCTAAGA......................................................................................... | 17 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .............................................................AGCGATTCCCGGTATGGGAG......................................................................................................................................................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................AATGGCTTAGTGAGGCC....................................................................................................................................................................................................................... | 17 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................GCCCCTACTCCGCCA.................................................................................... | 15 | 1 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................................................................................ACTGTGAACCTGCGAATGG..... | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
GCAAAGCGTGAGTTTCAATTACCGAACCAATCCGGTGCGGGACGAGTAGATCTTAAAGCTTTCGCTAAGGGCATTACCGTCGCTCGGTAATCGGAGTTCACCGTTACACCGTTTCAACGCAGATTTGAAAGGTAAACAAGGCAAATTCGACCGGGGATTAGGCGGTAGTTTATCATTGAAAGAGGCGAGAAGAAAGCGTCCAAAGAAGTTAGTTTCCCTTTTTAGTTGACAATTGGCCGCTCACCTGCGT
**************************************************.......((((((((.....))(((((((.....)))))))(((......)))...((((.((((((((((.(((((..............))))).))((((.........)))).......))))))))..))))....))))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
GSM343915 embryo |
SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
M053 female body |
|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................GAAAGCGTCATCAGAAGTTA....................................... | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................AAAGCGTTCTCTAAAGGCATT............................................................................................................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................TCCAAAGTAGTTAGGTT................................... | 17 | 2 | 17 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................AGCCGGTGCGGGATTAGTA.......................................................................................................................................................................................................... | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................AGGAAGGCGTCCAAAGATG.......................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 10:50 AM