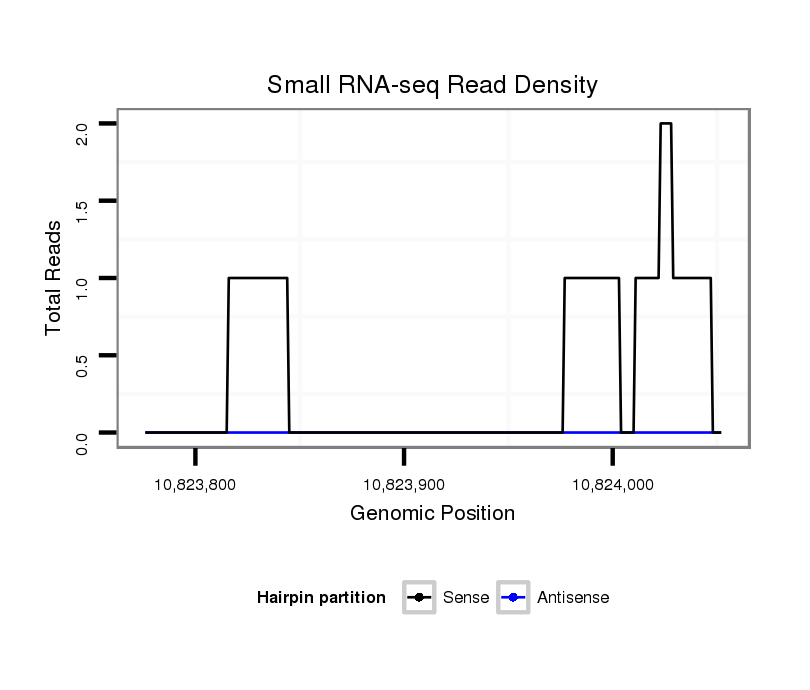
ID:dsi_30504 |
Coordinate:3r:10823826-10824002 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
five_prime_UTR [3r_10824034_10824040_+]; CDS [3r_10824041_10824100_+]; exon [3r_10824034_10824100_+]
No Repeatable elements found
| ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################### TGTGCAACAAAATTTTATTTGAGTACAAAATATGTTTAAATTTTAGATTCGCGTGTAAGATTTACGAGCTTCTATGACTTCCCAATTGTTTGGTTGTTTAGTTTTATTTTATAATTATCATGACGTCACATCTTACATCCAAGCGAAGCGAGAACAATGAGCGTGAATATTTGAAATTCATATTTCGCAATTAAACGTAGATGCGCTGACCAGCTCTGATTTTTCAGTTAGTTTCGCTTTTAGCCGATCAGCGAACGGTTGAGCCATGGATTTATTT **************************************************.((((.......)))).((.((((((.........((((((((.(((...........((((.......))))...........))).)))))))).((((((...(((((...............))))).)))))).......)))))).))(((....))).((((....))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M053 female body |
O001 Testis |
SRR553485 Chicharo_3 day-old ovaries |
SRR553486 Makindu_3 day-old ovaries |
GSM343915 embryo |
M024 male body |
M023 head |
SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR902009 testis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................................................TCAGCGAACGGTTGAGCCATGGATT..... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................GCTTTTAGCCGATCAGCG........................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................TTTTAGATTCGCGTGTAAGATTTACGAGC................................................................................................................................................................................................................ | 29 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................TGCGCTGACCAGCTCTGATTTTTCAGT................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................CGGTAGAGCAATGGATTGA... | 19 | 3 | 20 | 0.80 | 16 | 0 | 0 | 0 | 0 | 0 | 11 | 4 | 0 | 0 | 1 |
| ......................................................................................TGTTTGGTTATCTAGTTTTATG......................................................................................................................................................................... | 22 | 3 | 6 | 0.33 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................TCTTTGGTTATTTAGTTTTATGT........................................................................................................................................................................ | 23 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................TGTTTGGATTTTTAGTTTTATT......................................................................................................................................................................... | 22 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................TCTTTGGTTATTTAGTTGTATTT........................................................................................................................................................................ | 23 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................TTTGGTAGTTTAGTT.............................................................................................................................................................................. | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................GTGAATATTTGGAAT.................................................................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................................................................................................................ATCGGTAGAGCAATGGATT..... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
|
ACACGTTGTTTTAAAATAAACTCATGTTTTATACAAATTTAAAATCTAAGCGCACATTCTAAATGCTCGAAGATACTGAAGGGTTAACAAACCAACAAATCAAAATAAAATATTAATAGTACTGCAGTGTAGAATGTAGGTTCGCTTCGCTCTTGTTACTCGCACTTATAAACTTTAAGTATAAAGCGTTAATTTGCATCTACGCGACTGGTCGAGACTAAAAAGTCAATCAAAGCGAAAATCGGCTAGTCGCTTGCCAACTCGGTACCTAAATAAA
**************************************************.((((.......)))).((.((((((.........((((((((.(((...........((((.......))))...........))).)))))))).((((((...(((((...............))))).)))))).......)))))).))(((....))).((((....))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
SRR902009 testis |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|
| ...............................................................TGGTCGAAGAAACTGAAGGGGT................................................................................................................................................................................................ | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 |
| ...............................................................................................................................................................................................................CTGGTCGAGATTAAAA...................................................... | 16 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 |
| ................................................................................................................................................................................................................TGGTCGAGATTAAAAA..................................................... | 16 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 |
| .....................................................................CAGATACTGAAGGGT................................................................................................................................................................................................. | 15 | 1 | 6 | 0.17 | 1 | 0 | 0 | 1 |
| ...............................................................................................................................................................................................................CTGGCCGAGTGTAAAAAGTC.................................................. | 20 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 |
| ..........................................................................ACGGAAGGGTTAGCAAA.......................................................................................................................................................................................... | 17 | 2 | 14 | 0.07 | 1 | 0 | 0 | 1 |
| .....................................................................................................................................................................................................................GAGACTAAAATGTCAA................................................ | 16 | 1 | 17 | 0.06 | 1 | 1 | 0 | 0 |
Generated: 04/24/2015 at 10:50 AM