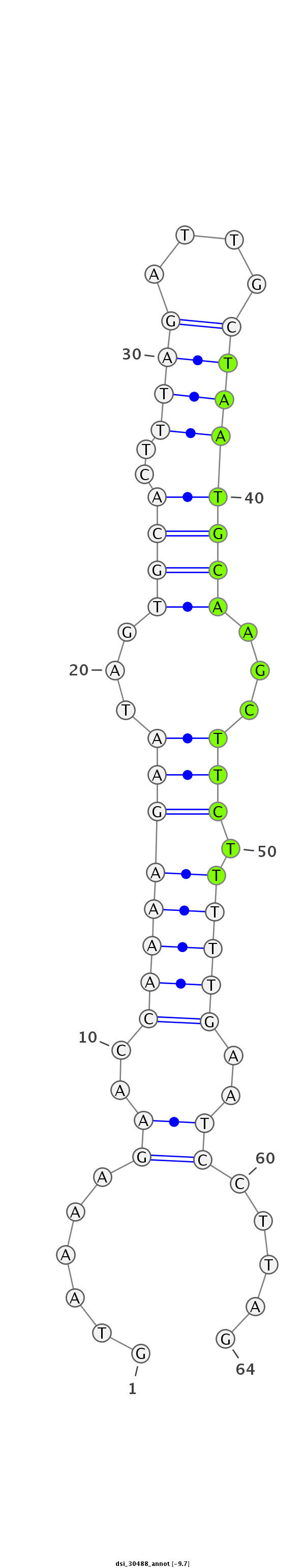
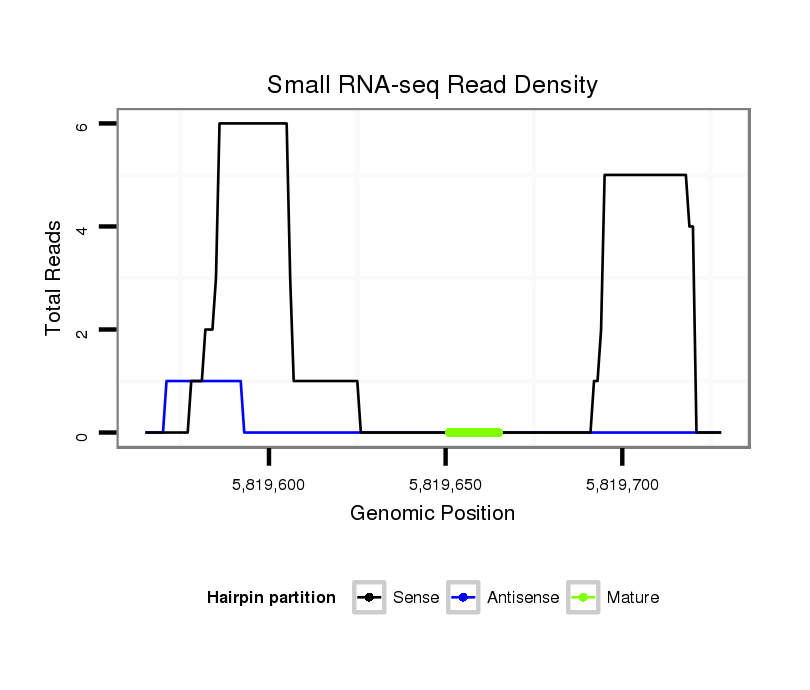
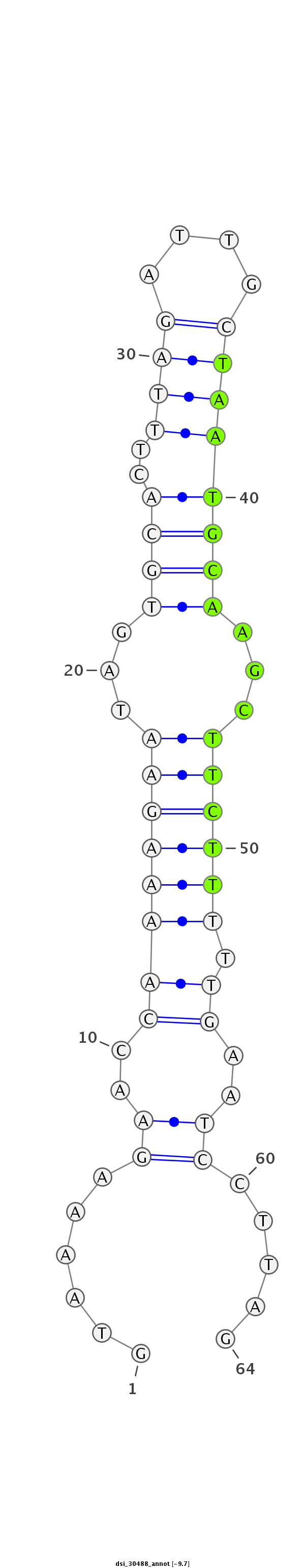
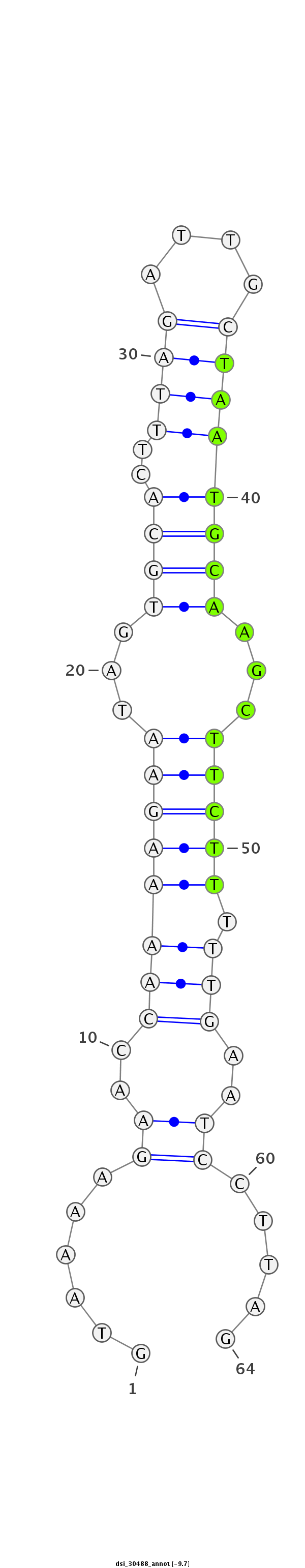
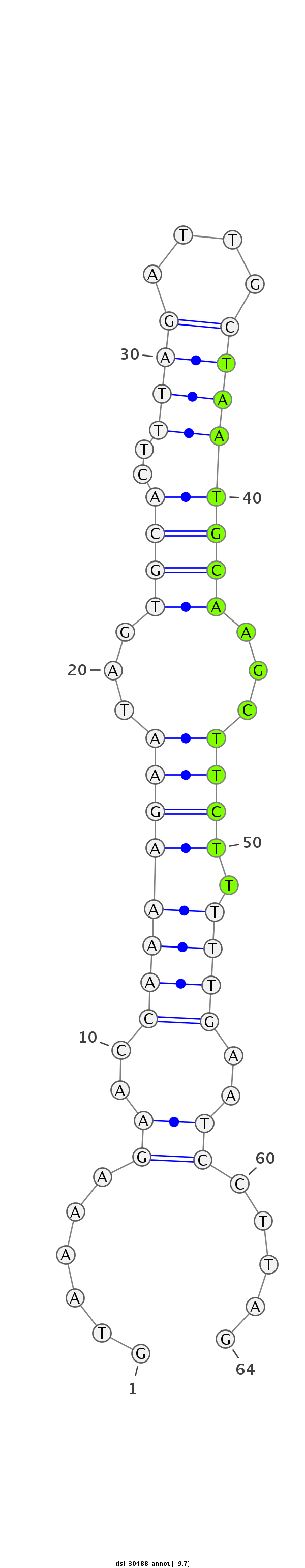
ID:dsi_30488 |
Coordinate:2r:5819615-5819678 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -9.7 | -9.7 | -9.7 |
 |
 |
 |
intron [2r_5819728_5819781_+]; CDS [2r_5819679_5819727_+]; exon [2r_5819679_5819727_+]; CDS [2r_5819520_5819614_+]; exon [2r_5819297_5819614_+]; intron [2r_5819615_5819678_+]
No Repeatable elements found
| mature | star |
| ##################################################----------------------------------------------------------------#################################################- AGGGCGTGCATCCTGGTAGCCAAACTGATTGGACTGGACCTGGAGCTCAAGTAAAAGAACCAAAAGAATAGTGCACTTTAGATTGCTAATGCAAGCTTCTTTTTGAATCCTTAGACCCGTTGACTTCGCCAAGAAGGAGCACCTGAGCGAGGAATTTGTCAAGG **************************************************......((..((((((((...((((..((((....))))))))...))).)))))..)).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
M025 embryo |
SRR553488 RT_0-2 hours eggs |
M023 head |
SRR553485 Chicharo_3 day-old ovaries |
SRR902008 ovaries |
GSM343915 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................AGCACCTGAGCGAGGAATTTGTCAAGC | 27 | 1 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................AAGAAGGAGCACCTGAGCGAGGAATT........ | 26 | 0 | 1 | 3.00 | 3 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................AAACTGATTGGACTGGACCTG.......................................................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...........................CTTGGACTGGACCTTTAGCTCA................................................................................................................... | 22 | 3 | 2 | 1.50 | 3 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................TCAAACCCGTTGACTTCGCCA................................. | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................GCCAAGAAGGAGCACCTGAGCGAGGAA.......... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................CAAACTGATTGGACTGGACCT........................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................CAAACTGATTGGACTGGACCG........................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................AGCCAAACTGATTGGACTGGACCT........................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................TGGAGCTCAAGTAAAAGAACC....................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................TGGACTGGACCTTTAGCTC.................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................AGCACCTGAGCGAGGAATTTGTAAAGC | 27 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................AAACTGATTGGACTGGACCT........................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................CAAGAAGGAGCACCTGAGCGAGGAATT........ | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............TGGTAGCCAAACTGATTGGACTGGACC............................................................................................................................ | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................TAATGCAAGCTTCTT............................................................... | 15 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................GATGCAAGCTTCTTT.............................................................. | 15 | 1 | 10 | 0.50 | 5 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................CTGATGCAAGCTTCTT............................................................... | 16 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................TGATGCAAGCTTCTTT.............................................................. | 16 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................GATGCAAGCTTCTTTC............................................................. | 16 | 2 | 20 | 0.15 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................GCTGATGCAAGCTTC................................................................. | 15 | 1 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................AGCTCGAGTAAAAGA.......................................................................................................... | 15 | 1 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................AGTCCCGTCGTCTTCGCCA................................. | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ..................................................................................TCGATGATGCAAGCTTCTT............................................................... | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................TGGTGATCAAGTAAAAGAA......................................................................................................... | 19 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................ACTGATGCAAGCTTCTT............................................................... | 17 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................GGTGATGCAAGCTTCT................................................................ | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................CGCGGATGCAAGCTTCTT............................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................CTTTGGATTGCTAAT.......................................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TCCCGCACGTAGGACCATCGGTTTGACTAACCTGACCTGGACCTCGAGTTCATTTTCTTGGTTTTCTTATCACGTGAAATCTAACGATTACGTTCGAAGAAAAACTTAGGAATCTGGGCAACTGAAGCGGTTCTTCCTCGTGGACTCGCTCCTTAAACAGTTCC
**************************************************......((..((((((((...((((..((((....))))))))...))).)))))..)).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR902009 testis |
SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
M053 female body |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ......ACGTAGGACCATCGGTTTGACT........................................................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................TCTAGGCAAATCAAGCGGTT................................ | 20 | 3 | 8 | 0.38 | 3 | 0 | 0 | 3 | 0 | 0 | 0 |
| .............................ACCTGTGCTGGACTTCGAGTT.................................................................................................................. | 21 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................TCGGATTGACGAACCTGAC................................................................................................................................ | 19 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................GTTAATGTTCTGGGTTTTCT................................................................................................. | 20 | 3 | 20 | 0.15 | 3 | 0 | 0 | 0 | 3 | 0 | 0 |
| .................TCGGATTGACGAACCTG.................................................................................................................................. | 17 | 2 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................CGAGTTCCTTTTCTAGGT...................................................................................................... | 18 | 2 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................GAATAACAAGACCTGGACC......................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 12:34 AM