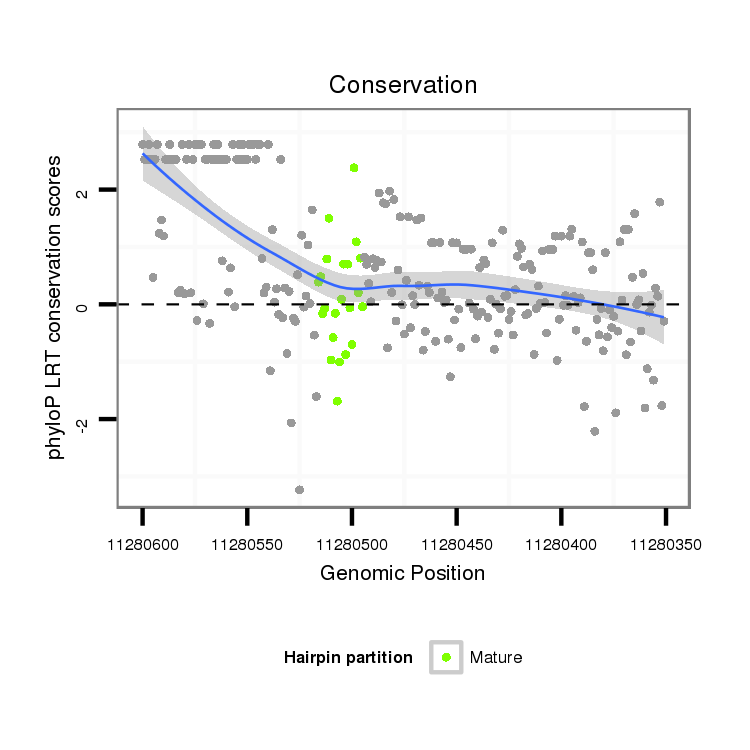
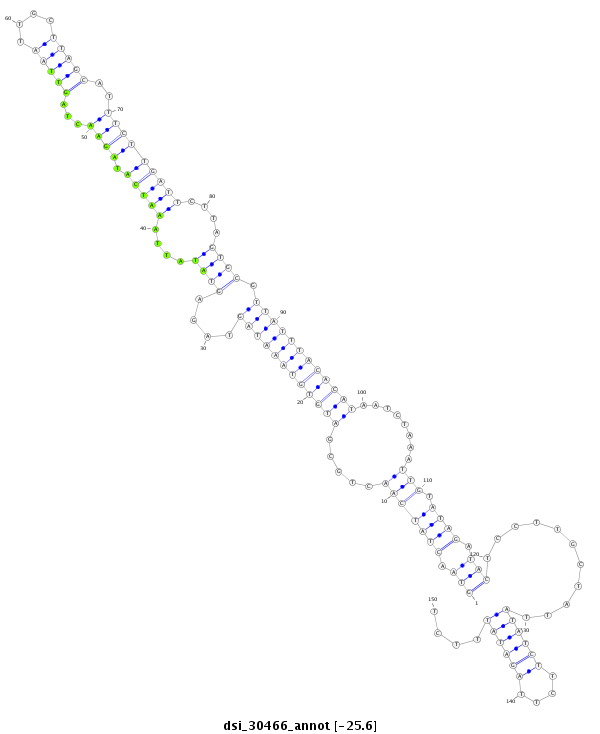
| droSim2 |
3r:11280351-11280600 - |
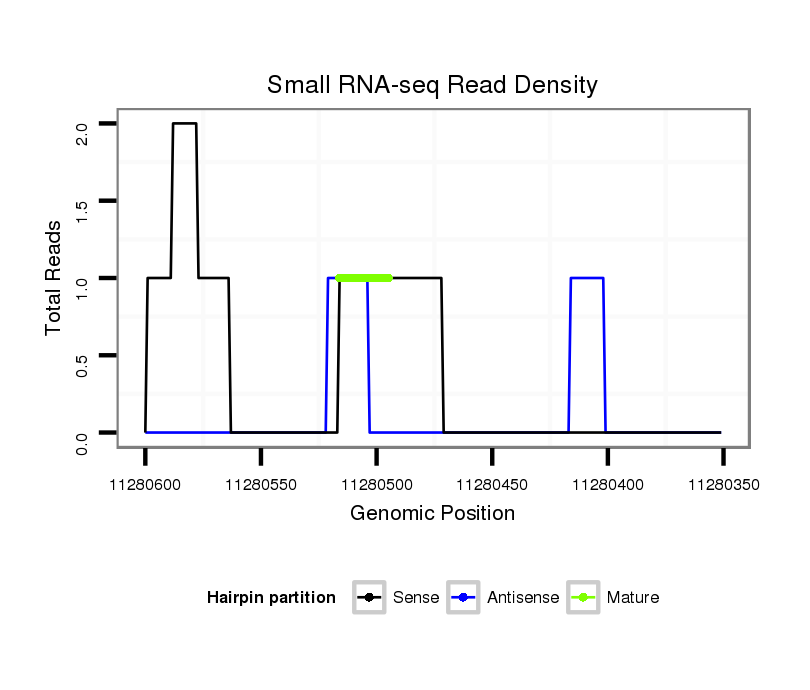
dsi_30466 |
AGCAGCGTCAGCGAGGGGCTGGACCACTTCCCCGAGAGTCCGCTCCACAGGTAACTATCAACTGCGAT---GTGTA-----------------------------AAT---AGTA--GAGTATATT-----------------------------------------AAATCATAG---AACTAGTTAAT--------------------------------------------------------TGCTTAGCATTT--CTTGATTCTTAGT-GCGTTATTTACACATAATCTAAATTG-----------TATAGATACTCCTTGC---------------------------------------------------------------------------------------------------------------------------------------------------------------------TAT-------TATATCTT--------------------------------------------------------------------------------CTTAGATATTTCTTAATTATTTTTTCATATAACAAG--------------------------------------------------------------------------------------------------------AACGAAA--------TAT------------------------AA------------AATCTG-----------------------GATC-TTAGG |
| droSec2 |
scaffold_0:12110704-12110953 - |
|
AGCAGCGTCAGCGAGGGGCTGGACCACTTCCCCGAGAGTCCGCTTCACAGGTAACTATCAAATGCGAT---GAGTA-----------------------------AAT---AGTA--GAGTATATT-----------------------------------------AAATCATAG---AACTAGTTAAT--------------------------------------------------------TGCTTAGCATTT--CTTGATTCTTAGT-GCGTTATTTACACATAATCTAACTTG-----------TATAGATACTCCTTGC---------------------------------------------------------------------------------------------------------------------------------------------------------------------TAT-------TATATCTT--------------------------------------------------------------------------------CTTAGATATTTCTTAATTATTTTTTCATAAAACAAG--------------------------------------------------------------------------------------------------------AACGAAA--------TAT------------------------AA------------AATCTG-----------------------GATC-TTAGG |
| dm3 |
chr3R:9905311-9905560 + |
|
AGCAGCGTCAGCGAGGGGCTGGATCACTTTCCCGAGAGTCCGCTCCACAGGTAACTATCAACTGCGAT---GAGTA-----------------------------AAT---AGTA--GAGTATATTAA---------------------------------------AAATCATAGAACAACTAGTTAAT--------------------------------------------------------TGCTTAGCATTT-TCTTGATTCTTAGT-GCGTTATTTACACATAATCTAACTTG-----------TATAGATACTCCTTGC---------------------------------------------------------------------------------------------------------------------------------------------------------------------TAT-------TATATCTT--------------------------------------------------------------------------------CGTAGATATTTCTTAATTATT---AAAT---ACAAG--------------------------------------------------------------------------------------------------------AACGAAA--------TAT------------------------AA------------AATCTG-----------------------GATC-ATAGG |
| droEre2 |
scaffold_4770:9445946-9446142 + |
|
AGCAGCGTCAGCGAGGGGCTGGACCACTTCCCCGAGAGTCCGCTCCACAGGTAACTATCAACTGCGAT---GAGTA-----------------------------GAC---AGTA--GAGCATT-----------------------------------------T-AAGCCATAG---AACTAGTTAAA--------------------------------------------------------TGCTTAG-------------------T-GCG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AG-------TATAAC----------------------------------------------T-----------------------------TAACCTTGTAGACATTTCTTAATTATATTTGCATACCTCAAA--------------------------------------------------------------------------------------------------------AACCGAT--------TTT------------------------GA------------AATCTG-----------------------GATC-TAAAG |
| droYak3 |
3R:16526149-16526400 - |
|
AGCAGCGTCAGCGAGGGGCTGGACCACTTTCCCGAGAGTCCGCTCCACAGGTAACTATCAACTGCGAT---GCGCA-----------------------------GAG---GGTA--GAGAATT--GG---------------------------------------AAATCATAG---AACTAGTTAAT--------------------------------------------------------TGCTTAGCATTT---TTGATTCTTAGT-GCGTTACCTACACGTAATATAACTTA----TGTA---TATAGATAT------A---------------------------------------------------------------------------------------------------------------------------------------------------------------------TGT-------TATATCTT--------------------------------------------------------------------------------CGTAGACATTTATTAATTATATTTTCATACAACCAA--------------------------------------------------------------------------------------------------------ACCAAAA-CCGAT--TAT------------------------AA------------AATCTG-----------------------AATT-TTAGG |
| droEug1 |
scf7180000409802:35851-36127 - |
|
AGCAGCGTCAGCGAGGGGCTAGACCATTTCCCGGAGAGTCCGCTGCACAGGTAACTATTAAATGCGAT---GAGTC-----------------------------AAGTACAGTA--A-----ACT---------------------ATCTATATATAAAGA---AAACTTACTAGAAAATATAGTTAAT--------------------------------------------------------TGCTTAGCCTTT-CATTACTTTTTAGT-AAGTTATTTAGCGGTT---------------------------------------------------------------------------------------TTAAT--TATTT-------------------TTCGAACTATTTATCTTTAACTA--------ATAATCCATTTT-------------------------------GTTAGCTAA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------TT-------GGCTAGTTAATATTTAATAAATTAATAAAC--------------TTTTTCAATAAA----------------------------------------------------------------T-----------------------AAAC-TTAGG |
| droBia1 |
scf7180000302402:2693572-2693821 + |
|
AGCAGCGTCAGCGAGGGGCTCGACCACTTCCCCGAGAGTCCGCTCCACAGGTAACTATCAACTGCAAT---GAGCT-----------------------------TTGTATAGTT--T--------------------------CTTATTT-----TAGAGTACTTAACTT------------------------------------------------------------------------------------------------ACTAGTT-AG---------------------------------------A---------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAGATAT-------T-----------------TTACCTTAG--------------CGCACA---CTTTTTTTAGTAACATTTTAAATAAAAAAA--------------------------ATATTTTCATAAAACTAAAAAGTTTATTAAAAAAAGGAAAGCTCT-----------------------GATA---------------------AGGGAAGCA------------GAAG----ATCAAA----------------------------------------------------ACAA-----------------------TCTT-ATAAT |
| droTak1 |
scf7180000415380:796876-797232 + |
|
AGCAGCGTCAGCGAGGGGCTGGACCATTTCCCCGAGAGTCCGCTCCACAGGTAACTATCAACTGCAAT---GCGAT-----------------------------ATATTCAATA--G--------------------------CCTTCCT-----TAGAGTATTTAACTTTCTTA---AACTAGTTAAT--------------------------------------------------------TGCTTAGCCTTT-TATTATTTCTAAGT-GCCTTACTTTGACTTTA----------------------AAGAAAGTCC------------------------GTATATTTGTTAAATGAAAAATGAGTG-CCTTACT--TACCTTAAAGAAAGTCCGTATATTTATTAGCTTTTTTTCTTAAGCAAACATTTTAAAAATCCTTTTTAATCAAGAAAAAAAATGTTAAAAAAAAATCAGTTGGTTGA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTCTTAGGTG-TCTCCTA-------------ACTGAGAGA---------------------A-------------------------------------------------A------------AATCTA-----------------------GA---TAACT |
| droEle1 |
scf7180000491047:1254704-1254993 - |
|
AGCAGCGTCAGCGAGGGGCTGGACCATTTCCCAGAGAGTCCGCTCCACAGGTAACTATCAACTGCGAT---GAGTA-----------------------------AAATAAAGTA--G-----ATT-----------------------------------------ATACAGTAT---AAGTAGTCAAA--------------------TAGCTTTAAAAGCCTTTCATTACTTCTTGGTTGAATTGCTTAGCCTTT-TATTACATCTTAGT-TAG---------------------------------------C---------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGAAATTATTAAAAA---AT---------TTTTACCTTGG--------------CTTACG---CT--------------------------------------------------------------------------------------------------------------------------T--A---------------------AGGAAAATATACAAATTTATAAAAA--------TAACCCA--------CT---------------------TA---AAA--GTAAAATATGGGGCATAAAATGTTAACTGTCAAGCTTTT---- |
| droRho1 |
scf7180000771044:15641-15955 + |
|
AGCAGCGTCAGCGAGGGGCTGGACCATTTCCCAGAGAGCCCGCTCCACAGGTAACTACCAACTGCGAT---GAGTA-----------------------------AACTGTAGTA--G-----ATT-----------------------------------------ATACGATAT---AACTAGTTAATAGCTTAG-------------------------ACTTTCATCGCCTCTTAGTTGAACTGCATAGCCTTT-CATTACTTCTTAGTT-GG---------------------------------------C---------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGGAAATTATTAAATATATATTTTTAGATATTTTATCTTAATAGTAATATTTAAGCTTA---------------------------------------------------------------------------------------------AAAGGAAAGTGCT-----------------------T------------------------AGGAAAATAAAAAAG--------------TATTTAAACTA--------TT---------------------TATTTAAA--GTAGAATTTTAGTCGTAA----TTGATTG-GAAGCTCATTAGG |
| droFic1 |
scf7180000453912:410966-411248 + |
|
AGCAGTGTCAGCGAGGGGCTGGACCACTTCCCGGAGAGTCCGCTCCACAGGTAACTATCAACTGCAAA---GAGAA-----------------------GTAACTAAATACAGTA--GAACATT-----------------------------------------T--AATC-------AACTAG---AT--------------------------------------------------------TGCTTAGCTTTTTTACTACTTCCTAGTTAAGT-----A-----AAACTAACTAATTTATGATTAAAATAGTTACGTCTTGCTTATGATTTTTAAAGATTTCTTATAT--------------------GACTTTATTAACACCTTAAAT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAA-------CTATGTATGT------------------------TT--TAATTTGT--AGTTTCAATTCCAA-----TGATTC-GAAGTTC-TTAAA |
| droKik1 |
scf7180000302475:1231924-1232109 - |
|
AGCAGTGTATCCGAGGGATTGGATCATTTTCCGGAGAGTCCGCTGCACAGGTAACTACCAACTGCGAG---TAGAA-----------------------GGCTTAGATTAGAGTA--G-----A----------------------------------------------------------TAATTTAT--------------------------------------------------------TGCTTAGCTTG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAG-------T-----------------TA-------------------------------TT---------------------------------------------TTATTAATAAG---AAAT---TAAAG--------------------------------------------------------------------------------------------------------TATAAAA--------TATATTTTTTTGTTTAGAAATATCGTTAA------------AGTTTG-----------------------CATA-T---- |
| droAna3 |
scaffold_13340:20570270-20570499 - |
|
AGCAGTGTCAGCGAGGGGCTGGACCATTTTCCAGAGAGCCCGCTGCACAGGTAACTATTAATTTTTATTATTTTCCAGCTTTTTAATGAATTTTTTGAAAAGTTA---------------------------------------------------------------AATTAGAA---ATATAGTAAAT--------------------------------------------------------AA-------TTT--CT------TAAAT-AAGATATTTAGAATTAAAATAACTA-----------------ATACTAGTCAC---------------------------------------------------------------------------------------------------------------------------------------------------------------------TTT-------TAT----------------------------------TAAGTTTT---------------------------------------------------------------------------------------------AAAGGCATGTTGTTTATTTAGTTT-------GATTAGTTA---------------------A-----------------------------------------------------------------------------------------A-----------------------GAAT-TTAGA |
| droBip1 |
scf7180000396413:1832652-1832796 + |
|
AGCAGTGTCAGCGAGGGGCTGGATCATTTTCCAGAGAGTCCCCTGCACAGGTAACTATTTAATTTGAT---TTTTA-----------------------------------A---------------------------------------------------------------------------------------------------------------------------------------------CATTT-TA----------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTAATTTTAATTAAA--------------------------------------------------CT-------T----------------------------------------------TTTGAT-----------------AAATAAAGTAATAACTTTAGC-----------------------------------------------------------------------------TGATTTTTTA-------------AATGAG----------------------------------------------------------------------------------------------------------------------------AG |
| dp5 |
2:13484355-13484466 - |
|
AGCAGTGTCAGCGAGGGGCTGGATCACTTTCCGGAGAGTCCGCTGCACAGGTAACTATCAAATGTGAC---TCTCC-----------------------------AAA---AGGA--GACTCTT--CG-------------------------------------------TTTAC---AATTAGTTAGT--------------------------------------------------------CGCTTAG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droPer2 |
scaffold_0:6964828-6964938 + |
|
AGCAGTGTCAGCGAGGGGCTGGATCACTTTCCGGAGAGTCCGCTGCACAGGTAACTATCAAATGTGAC---TCTCC-----------------------------AAA---AGGA--GACTCTT--CG-------------------------------------------TTTAC---AATTAGTTAGT--------------------------------------------------------CGCTTA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droWil2 |
scf2_1100000004921:2062843-2063058 + |
|
AGCAGTGTCAGCGAGGGATTGGATCATTTCCCAGAGAGTCCATTACACAGGTAACTATTAAATTTAAA---CATAA-----------------------------------ATGATAAACACTT--AGTAAATAGTTAATGAGC-------------A-------C-AGGCCAGAA---CTTTAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAAATATTTGAAGAA--------------------------------------------------CT-------T----------------------------------------------TTTGTT-----------------------------GGCCTGTGT-----------------------------------------------------------------------------TGAATTTTTA-------------GAACAGGC--------------TTTTGCAAAAATAAAA-------CCACACATTT------------------------TA------------AAACTA-----------------------TAAT-TTAAG |
| droVir3 |
scaffold_12822:505908-505969 - |
|
AGCAGCGTCAGCGAGGGGCTGGATCACTTTCCAGAGAGTCCGCTGCACAGGTAACTATTTAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droMoj3 |
scaffold_6540:33178308-33178511 - |
|
AGCAGCGTCAGCGAGGGGCTCGATCACTTTCCAGAGAGTCCACTGCACAGGTAACTATTGAGGCCGAC---CTGAA-----------------------------------ACCA--GATCCAT----------------------------------------------------------TGAACAGTAGCCTAGATTTTGAATTAAG-----------------AATTCAGTTTAAGT-----AGTTTTGCATAT-TTTTTATTAT------------------------------------------------------TTGA---------------------------------------------------------------------------------------------------------------------------------------------------------------------TAT-------T---------------------------------------------------------------ATCTGGAATTACAGCAATATCCT-CATTGGTATTTAGTATTTATAGCTA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| droGri2 |
scaffold_15074:3272128-3272190 + |
|
AGCAGCGTCAGCGAGGGCTTGGATCACTTTCCAGAGAGTCCGTTGCACAGGTAACTATTAACT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |