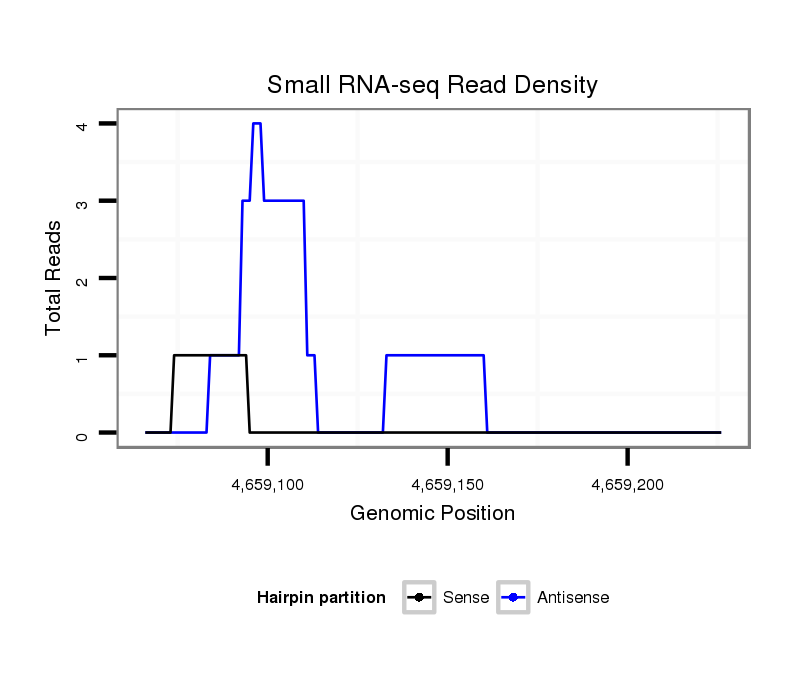
ID:dsi_30398 |
Coordinate:3r:4659116-4659176 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
exon [3r_4659002_4659115_+]; CDS [3r_4659002_4659115_+]; CDS [3r_4659177_4659397_+]; exon [3r_4659177_4659397_+]; intron [3r_4659116_4659176_+]
No Repeatable elements found
| ##################################################-------------------------------------------------------------################################################## AGTACCAGCAACAGGTCCACTCAGCGCCCGGTGTGCCGCCCCAGCAATACGTGAGTGCAAAAGCGCATTTCCGAACGGTAGACAATTAACTAGTAGGCCTCCGACTTCCAGCAATACGAACAGGTTCAAGTTCCCGTGCAGCAGCAGGCACCGGTCCAATA **************************************************..((((((......)))))).....(((.((.................)))))........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
GSM343915 embryo |
SRR618934 dsim w501 ovaries |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|
| ..........................CCCGGTATGTCGCCCCA...................................................................................................................... | 17 | 2 | 3 | 1.33 | 4 | 4 | 0 | 0 | 0 |
| ........CAACAGGTCCACTCAGCGCCC.................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ...........................CCGGTATGTCGCCCCA...................................................................................................................... | 16 | 2 | 13 | 0.46 | 6 | 6 | 0 | 0 | 0 |
| .........................TCCCGGTATGTCGCCCCA...................................................................................................................... | 18 | 3 | 20 | 0.45 | 9 | 9 | 0 | 0 | 0 |
| ............................................................................................................................................GCTGCCGGCACCTGTCCAAT. | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................CCCGTGGAGCAGAAGGCA........... | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 |
| .............................................................................................................AGCAATACGAAAAGGATC.................................. | 18 | 2 | 16 | 0.06 | 1 | 0 | 0 | 1 | 0 |
| ..............................................................................................AGGCCTCCGACGTCCG................................................... | 16 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
|
TCATGGTCGTTGTCCAGGTGAGTCGCGGGCCACACGGCGGGGTCGTTATGCACTCACGTTTTCGCGTAAAGGCTTGCCATCTGTTAATTGATCATCCGGAGGCTGAAGGTCGTTATGCTTGTCCAAGTTCAAGGGCACGTCGTCGTCCGTGGCCAGGTTAT
**************************************************..((((((......)))))).....(((.((.................)))))........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
M053 female body |
M023 head |
O001 Testis |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR1275487 Male larvae |
SRR902008 ovaries |
M025 embryo |
SRR902009 testis |
SRR553487 NRT_0-2 hours eggs |
O002 Head |
SRR553485 Chicharo_3 day-old ovaries |
GSM343915 embryo |
SRR1275483 Male prepupae |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................CACGGCGGGGTCGGTTTG............................................................................................................... | 18 | 2 | 10 | 115.50 | 1155 | 774 | 198 | 149 | 4 | 0 | 15 | 0 | 4 | 0 | 8 | 1 | 1 | 1 | 0 | 0 | 0 |
| ...............................ACACGGCGGGGTCGGTTTG............................................................................................................... | 19 | 2 | 6 | 20.17 | 121 | 51 | 45 | 5 | 7 | 1 | 3 | 1 | 4 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 |
| .............................CTACACGGCGGGGTCGGT.................................................................................................................. | 18 | 2 | 17 | 2.71 | 46 | 3 | 5 | 0 | 34 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 |
| ...............................ACACGGCGGGGTCCTGATG............................................................................................................... | 19 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................GGCCACACGGCGGGGTCG.................................................................................................................... | 18 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................TGAGTCGCGGGCCAC................................................................................................................................ | 15 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........GTCCAGGTGAGTCGCAGG.................................................................................................................................... | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................TGAGTCGCGTGCCAC................................................................................................................................ | 15 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................ACGGCGGGGTCGGTTTG............................................................................................................... | 17 | 2 | 20 | 1.00 | 20 | 0 | 0 | 0 | 0 | 0 | 12 | 4 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 1 |
| ..............................CACACGGCGGGGTCGTTA................................................................................................................. | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................AAAGGCTTGCCATCTGTTAATTGATCAT.................................................................. | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................GTCTACACGGCGGGGTCG.................................................................................................................... | 18 | 2 | 9 | 0.56 | 5 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................ACACGGCGGGGTCGGT.................................................................................................................. | 16 | 1 | 8 | 0.50 | 4 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................CTGAGTCGCGGGCCAC................................................................................................................................ | 16 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................CACGGCGGGGTCGGTTTGC.............................................................................................................. | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................ACACGGCGGGGTCGGTCT................................................................................................................ | 18 | 2 | 7 | 0.43 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................TCTGAGTCGCGGGCCAC................................................................................................................................ | 17 | 2 | 8 | 0.38 | 3 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................TCTGAGTCGCGGGCC.................................................................................................................................. | 15 | 2 | 20 | 0.30 | 6 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................CACGGCGGGGTCGGT.................................................................................................................. | 15 | 1 | 17 | 0.29 | 5 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ................................CACGGCGGGGTCGATTTG............................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................ACACGGCGGGGTCGTTTT................................................................................................................ | 18 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................TACACGGCGGGGTCG.................................................................................................................... | 15 | 1 | 20 | 0.20 | 4 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................TGAGTCGCGGGCCCCCC.............................................................................................................................. | 17 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................TGAGTCGCGGGCCACGG.............................................................................................................................. | 17 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................GCTACACGGCGGGGTCGGTTTG............................................................................................................... | 22 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................CACGGCGGGGTCGGTCTG............................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................GGTGAGTCGCGGGTTACA............................................................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................ACACGGCGGGGTCGCTTT................................................................................................................ | 18 | 2 | 15 | 0.13 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................GCTACACGGCGGGGTCGGTTT................................................................................................................ | 21 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................GCTACACGGCGGGGTCGG................................................................................................................... | 18 | 2 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................CACGGCGGGGTCGGTGTG............................................................................................................... | 18 | 2 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................TCTGAGTCGCGGGCCA................................................................................................................................. | 16 | 2 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................CGGGGTCGTTCTGGACTGA......................................................................................................... | 19 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................AGGCTGGGAATCTGTTAAT......................................................................... | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................CACGGCAGGGTCGGTTTG............................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................CCACGGCGGGGTCGGTTTG............................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................TGTCTACACGGCGGGGTCG.................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Generated: 04/24/2015 at 10:20 AM