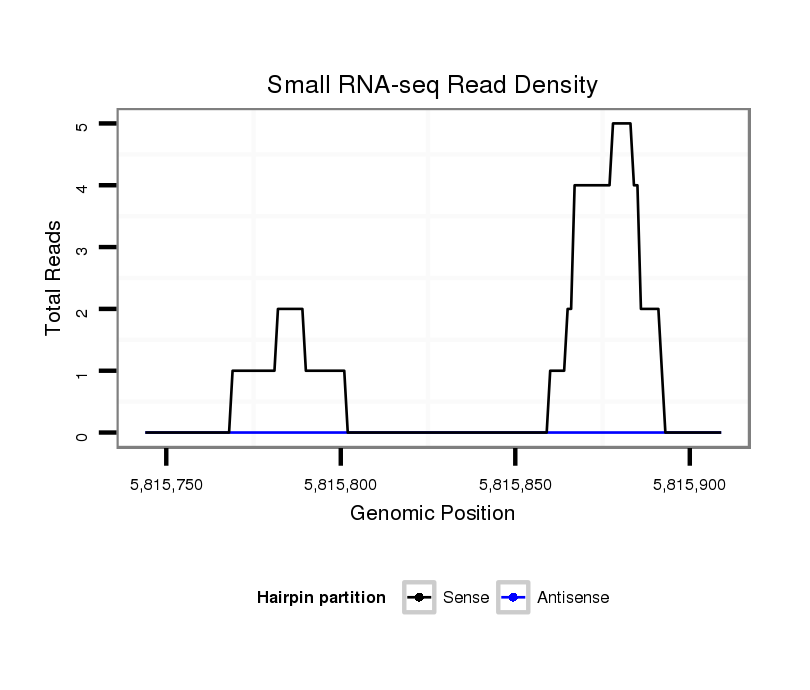
ID:dsi_30362 |
Coordinate:2r:5815794-5815859 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
intergenic
No Repeatable elements found
|
TACGCCTTCAAAGCCCGCCCAAAAGGTTGAGGTTATATCGGATGACATAGGTAATCGCTCCACACGCAAACTTTTAAATTGTTTTGCCATTAATCAAGTTTTTCGGCCATTATCAGAACTTATCTCGGACGACGCCAAGCCAGTAAAGAGGGATTTGTCCCCAGCA
**************************************************(((((.(((....((((((((.........)).))))..........)).....))).)))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
M025 embryo |
SRR553488 RT_0-2 hours eggs |
M024 male body |
SRR618934 dsim w501 ovaries |
SRR553487 NRT_0-2 hours eggs |
O002 Head |
SRR902008 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................TGAGGTTATATCGGATGACATAGAAC................................................................................................................. | 26 | 3 | 1 | 6.00 | 6 | 3 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................TAGGACGTCGCCAAGC.......................... | 16 | 2 | 16 | 3.19 | 51 | 29 | 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................TTGAGGTTATATCGGATGACATAGAAC................................................................................................................. | 27 | 3 | 1 | 3.00 | 3 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................CTCGGACGACGCCAAGCCA........................ | 19 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................CCAAGCCAGTAAAGA................. | 15 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................GTTGAGGTTATATCGGATGAC........................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................ATCTCGGACGACGCCAAGCCAGTAAAG.................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................CATAGAACTTATCTCGGACGACGCCA............................. | 26 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................ATATCGGATGACATAGGGAATCGCTC.......................................................................................................... | 26 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................AACTTATCTCGGACGACGCCAAGC.......................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................CGGATGACATAGGTAATCGC............................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................CAGAACTTATCACGGACC................................... | 18 | 2 | 6 | 0.33 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ...............................GTTATATCGTATACCATAGG................................................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............CCGCCCATATGGTTGAGG...................................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................CTAGGACGTCGCCAAGC.......................... | 17 | 2 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................GCCAAGCTAGTAAAGG................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................ATATCGGATGACATAGAAC................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................TGATATCGGATGACGT...................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................TCGGTCGCCGCCAAGCCT........................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
ATGCGGAAGTTTCGGGCGGGTTTTCCAACTCCAATATAGCCTACTGTATCCATTAGCGAGGTGTGCGTTTGAAAATTTAACAAAACGGTAATTAGTTCAAAAAGCCGGTAATAGTCTTGAATAGAGCCTGCTGCGGTTCGGTCATTTCTCCCTAAACAGGGGTCGT
**************************************************(((((.(((....((((((((.........)).))))..........)).....))).)))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
M024 male body |
SRR553486 Makindu_3 day-old ovaries |
M053 female body |
SRR618934 dsim w501 ovaries |
M023 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................AGCGCTGGGTGCGTTTGAAAA........................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....CGAAGTTTCGGGCTGGTT................................................................................................................................................ | 18 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....AGAAGTTTCGGGCTGGT................................................................................................................................................. | 17 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................AGAGCCTGCTGCGTT............................. | 15 | 1 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................ACTCTAACCAGTAGCGAGGT........................................................................................................ | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................TGAATTGAGCCTGCT.................................. | 15 | 1 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................CGGGTATTCCAAGTCCAA.................................................................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....CGAAGTTTCGGGCTGGTTA............................................................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 1 | 0 | 0 | 0 | 1 |
| ......AAGTTTCGGATGGGTTT............................................................................................................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....GGAAGTTTCGGACTGGT................................................................................................................................................. | 17 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
Generated: 04/24/2015 at 12:19 AM