| droSim2 |
2r:6989960-6990341 - |
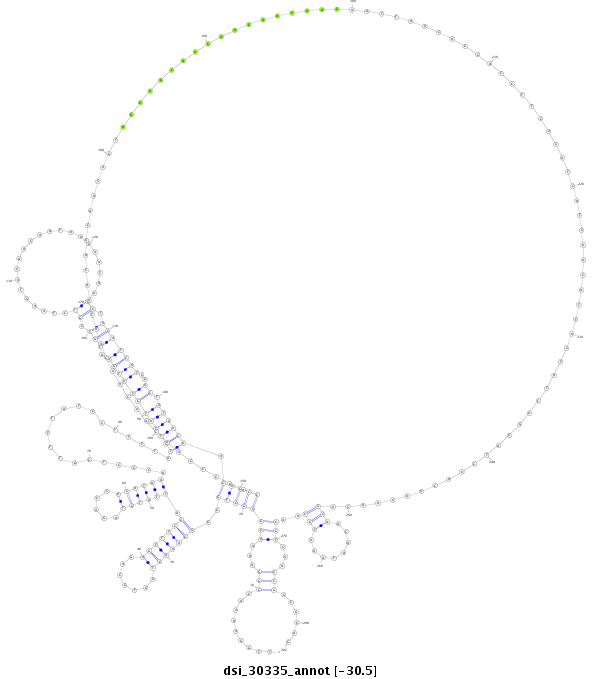
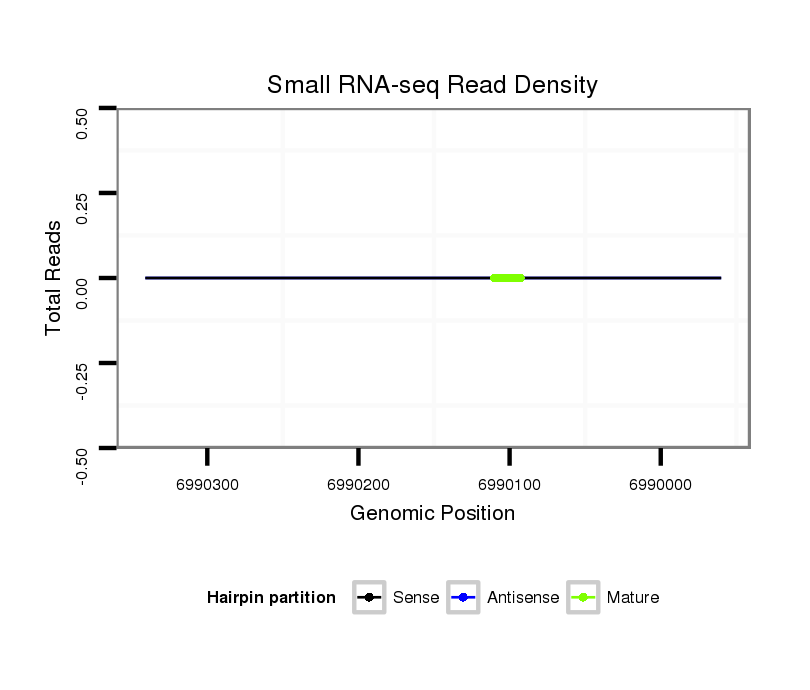
dsi_30335 |
GAGA---CTGGCTTG--------------G----AATTTAAGAATATAT---------------------CATCAATG-----T-----AAA--------------------------TTTGAT-CGAATTGG----------AAAACCGG-AAAAG-GG-----------------------------------------------------------------GCTG--GGG---------------------------------------------------------------------GAAACT----------------------A----GT-GCAG-AGTTTCCATT-------------------------------ATGTAGA-CCACAT------------AAAC-C----CTCGTTTTATCCTTTTATGTTATG----TATGAATGTCGTCTAAGTACAACAATAAC-----AATAA----------------A--------------------------CTACATCATC-GACTATGACA--ACAGCCAACCCAAC---------------------AAC------AACAAT---AACAACAACA------------------------------GTACAAGCAACAA------------CAACAACAATTAGCACCACCTTCACATCATCAACATTACTATCAGCATCAACAACAACAGCAACAATAAATGCAACCT------CA----------------------------AC------------CGCCGCAGCCGCACCACCAC---------------------------CAGTGCCGTCTGTTTGTGTGCGTGTGCCCCACA-------GTCCC |
| droSec2 |
scaffold_1:3828107-3828481 - |
|
GAGA---CTGGCTTG--------------G----AATTTAAGAA----TAAGAAAG--------------AATCAATG-----T-----AAA--------------------------TTTGAT-CTAATTGG----------AAAACCGG-AAAAGGGG-----------------------------------------------------------------GCTG--GGG-----------------------------------------------------------------------------------------------------------AG-AGTTTCCACT-------------------------------ATGTAGA-CCACAT------------AAAC-C----GTCGTTTTATCCTTTTATGTTATG----TGTGAATGTCGTCTAAGTACAACAATAAC-----AATAA----------------A--------------------------CTACATCATC-GACTATGACA--TCAGCCAACCCGAC---------------------AAC------AACAAT---AACAACAACA------------------------------GTACAAGCAACAA------------CAACAACAATTAGCACCACCTTCACATCATCAACATTACTATCAGCATCAACAACAACAGCAACAATAAATGCAACCT------CA----------------------------AC------------CGCCGCAGCCGCACCACCAC---------------------------CAGTGCCGTCTGTTTGTGTGCGTGTGCCCCACA-------GTCCC |
| dm3 |
chr2R:6221268-6221631 - |
|
ATGA---TT-----------------------------CAAGAA----T---------------------CATCAATG-----T-----AAG--------------------------CTTGAT-CGACTTGG----------AAAAGCGG-AAGAG-GG-----------------------------------------------------------------GCTG--GGG---------------------------------------------------------------------GAAACT----------------------A----GT-GCAG-AGTTTCCATT-------------------------------ATGTAGA-CCACAT------------AAAC-C----CTAGTTTTATCCTTTTATGTTATG----TATGAATGTC---TAAGTACAACAATAAC-----AATAA----------------A--------------------------CTACATCATC-GACTATGACA--ACAGCCAACCCAAC---------------------AAC------AACAAT---AACAACAACA------------------------------GTACAAGCAACAA------------CAACAACAATTAGCACCACCTTCACATCATCAACATTACTATCAGCATCAACAACAACAGCAACAATAAATGCAACCT------CA----------------------------AC------------CGCCGCAGCCGCACCACCAC---------------------------CAGTGCCGTCTGTTTGTGTGCGTGTGCCCCACA-------GTCCC |
| droEre2 |
scaffold_4929:8153229-8153610 + |
|
GAGA---TTGGCTTG--------------G----AAG-------TAGAT---------------------CATTAATC-----T-----AAA--------------------------CTTGAT-CAAATCGG----------AAAACCGG-AAAAG-GG-----------------------------------------------------------------GCTG--GGG---------------------------------------------------------------------GAAACT----------------------A----GT-ACA----------CT-------------------------------ATGTAGC-CCACAT------------AAAT-C----CTCGTTTTATCGTTTAATGTTATG----AATGAATGTCGTC--AGTACAACAATAAC-----AATAA----------------A--------------------------CTACATCATC-GACTATGACA--ACAGCCAACGCAACAAAAACAAC---------------------AACAAT------------AATAACAACAACAGCTAC------AACAACAGTACAAGCAACA---------------ACAACAATTAGCACCACCTTCACATCATCAACATTACTATCAGCATCAACAACAACAGCAACAATAAATGCAACCT------CA----------------------------AC------------CGCCGCAGCCGCACCACCAC---------------------------CAGTGCCGTCTGTTTGTGTGCGTGTGCCCCACA-------GTCCC |
| droYak3 |
2L:18846156-18846518 - |
|
ACTT---AA--------------------------------AAATGTAT---------------------CATTAATG-----T-----AAA--------------------------CTTGAT-CGAATTGG----------AAAATCAG-AAAGG-TG-----------------------------------------------------------------GCTG--GGG---------------------------------------------------------------------GAA-CT----------------------A----GT-GCAG-AGT---------------------------------------TGTAGA-AAACAT------------AAGCCC----CTTGTTTTATCGTTTAATGTTATG----TATGAATGTCGTCTAAGTACAACATTAAC-----AATAA----------------A--------------------------CTACTTCATC-GACTATGACA--ACAGCCAACCCAACAAAAACAAC---------------------AACAGTAACAACAACAACA------------------------------GTACAAGCAA------------------CAACAATTAGCACCACCTTCACATCATCAACATTACTATCAGCATCAACAACAACAGCAACAATAAATGCAACCT------CA----------------------------AC------------CGCCGCAGCCGCACCACCAC---------------------------CAGTGCCGTCTGTTTGTGTCCGTGTGCCCCACA-------GTCCC |
| droEug1 |
scf7180000409462:3665146-3665531 + |
|
GATA---ATAGATTTAACAC-------CAGA---ACTCT-------------------------------------TG-----C-----ACA--------------------------TTTGAT-TGAACTCG----------GAAA-TAGTAAACATGG-----------------------------------------------------------------GTTA-AGGG---------------------------------------------------------------------GAAACC----------------------T----CC-ACAA-AGTCTCCACA------------------------------CATGTAGT-CCACAT------------AAAC-A----CTCATTTGATCGTTAAATGTAATG----TATGAATGTCGCCTAAGTACAACAATATA-----AGTAA----------------A-C----------------------TACTACATCATC-AACTATGACA--ACAGCCAATCCAAC---------------------AAT------AA------CAACAACAACA------------------------------GTACAAGCAACAA---TAACAACAACAACAACAATTAGCACCACCTTCACATCATCAACATTACTATCAGCATCAACAACAACAGCAACAATAAATGCAACCT------CA----------------------------AC------------CGCCGCAGCCGCACCACCAC---------------------------CAGTGCCGTCTGTTTGTGTGCGTGTGCCCCACA-------GTCCC |
| droBia1 |
scf7180000301754:100531-100924 + |
|
ATAT---TTTATTTG--------------A----AGTCTTAAAA----T---------------------CA------------------TA--------------------------TTTGATTCGAATTCGATCAAAAAAAGAAAACAG-AAAAG-GG-----------------------------------------------------------------GCCGAAGGG---------------------------------------------------------------------GAAACC----------------------TA---TC-TCAT-AGTTTCCTCT-------------------------------GAACACA-CCACAT------------AAAC-A----CTAGTTTGATCGTTTAATGTTATG----TATGAATGTCGCCTAAGTACAACAATGAA-----AGTAA----------------A-------------------------ACTGCATCATC-AACTACGACA--ACAGCCAATCCAACAACAACAAC---------------------AACAGT---AACAACAACA------------------------------GTACAAGCAACAA---------CAACTACAACAATTAGCACAACCTTCACATCATCAACATTACTATCAGCATCAACAACAACAGCAACAATAAATGCAACCT------CA----------------------------AC------------CGCCGCAGCCGCACCACCAC---------------------------CAGTGCCGTCTGTTTGTGTGCGTGTGCCCCTCA-------GTCCC |
| droTak1 |
scf7180000415683:256806-257180 + |
|
GAGATGTTTTATTCA--------------G----AATTTTGAAA----TAAGATATAAAGGATTTTATATT----------GCT-----TAA--------------------------TTTGAT-CGAAATTG----------GAAA-TA---AAAGAGG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GT-CCACAT------------AAAC-ATAGACTAGCTTGTTCCTTTAATGTTATG----TATGAATGTCGCCTAAGTACAACAATAATGTTTAAAAAA----------------A--------------------------ATACATCATC-AACTATGACA--ACAGCCAATCCAAC---------------------AAC------AACAATAACAACAGCAACA------------------------------GTACAAGCAACAA---------CAACAACAACAATTAGCACAACCTTCACATCATCAACATTACTATCAGCATCAACAACAACAGCAACAATAAATGCAACCT------CA----------------------------AC------------CGCGGCAGCCGCACCACCAC---------------------------CAGTGCCGTCTGTTTGTGTGCGTGTGCCCCACA-------GTCCC |
| droEle1 |
scf7180000491057:27541-27944 - |
|
AAGA---ATTGTTTTAAG-C-------CAAAATGA--------ATATAT-----ATAAAACAT---ATATTGTA-------GCT-----AAAAC------------------------TTTGAT-CGGATTTG----------GAAA-CAG-AGAGG-AG-----------------------------------------------------------------TGTA--AGG---------------------------------------------------------------------GAAATC----------------------G----CT-GCAG-GATTTCTATT-------------------------------GTGTAGTCCCACAT------------AAAC-A----CTCGTTTGATCGTTTTATGTAATG----TGTGAATGTCGCCTAAGTACAAGAATAAC-----AAACA----------------AAA----------------------AACTACATCATC-AACTATGACA--ACAGCAAATCCAAC---------------------AAT------AA------CAACAACAACA------------------------------GTACAAGCAACAA------CAACAACAACAACAATTAGCACAACCTTCACATCATCAACATTACTATCAGCATCAACAACAACAGCAACAATAAATGCAACCT------CA----------------------------AC------------CGCCGCAGCCGCACCACCAC---------------------------CAGTGCCGTCTGTTTGTGTGCGTGTGCCCCACA-------GTCCC |
| droRho1 |
scf7180000769355:16203-16589 + |
|
AAAA---C-----------------------------------------------------AT---ACATTGGTTATAATAGCTTTCTTAAGGCTTTATCAGCTATACATCCGCTACTTTTGAT-CAAATTCG----------AAAA-CAG-AAAGG-GG-----------------------------------------------------------------GTTA--AGG---------------------------------------------------------------------GAAAGC----------------------C----CT-GCAG-G----------------------------------------ATGTAGTCCCACGT------------AAAC-A----CTCGTTTGATCGTTTAATGTAATG----TGTGAATGTCGCCTAAGTACAACAATTAC-----AATCA----------------AAA----------------------AACTACATCATC-AACTATGACA--ACAGCAAATCCAAC---------------------AA------------------CAACAACA------------------------------GTACAAGCAACAA------------CAACAACAATTAGCACAACCTTCACATCATCAACATTACTATCAGCATCAACAACAACAGCTACAATAAATGCAACCT------CA----------------------------AC------------CGCCGCAGCTGCACCACCAC---------------------------CAGTGCCGTCTGTCTGTGTGCGTGTGCCCCACA-------GTCCC |
| droFic1 |
scf7180000454066:1487266-1487700 + |
|
AACC---TTG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTG--GGGGATTAAATAAGATAATATTTATAACTGTTCATAATTGAAAGTTTATTATTGCCATATTGAATCGGCTTAGAAATCGGTGTAGGGCATAGGAAAAATCT----TT-AAAG-AGTTTTTACT-------------------------------CCGTAGT-ACACAT------------AAAC-A----CTTGTTTGATCGTTAAATGTAATG----TGTGAATGTCGCCTAAGTACAACAATAAC-----AAAAACCAAAAAAAAAAAATAA--------------------------CTGAATCATC-AACTATGACA--ACAGCAAATCCAAC---------------------AAT------AA------CAACAACAACA------------------------------GTACAAGCAACAATAACAACAACAACAACAACAATTAGCACCACCTTCACATCATCAACATTACTATCAGCATCAACAACAACAGCAACAATAAATGCAACCT------CA----------------------------AC------------CGCCGCAGCCGCACCACCAC---------------------------CAGTGCCGTCTGTTTGTGTGCGTGTGCCCCACA-------GTCCC |
| droKik1 |
scf7180000302817:558343-558777 - |
|
AGGA---CTC-------------------------------------------------------------------------------------------------------------------------------------------------AGGGCACTCAGGAGTCTTTCTTTAACAAATCCAAATACCATTCAGTTCAAAACTCTTAAATCGAAATCCGACTG--AAA---------------------------------------------------------------------GAAATTATTTTAGGGTG-----------A-CTCCTGACAGTAGCTT--------------------------------------------CCAAATAAAATTAAAAAAAAAC-A----CTTGCAATATTGTTTAAAGTAATGTTTGTGTGAATGTCT--TAAATACAA--ATAAC-----AAAA----------------------------------------------TCATCATCCAACTATGACAAAACAGCTATTCCAACAACTACAACAACTACTC---------------CAATAACAACAACAAC------------AACTCCAATTAAAACAACAATACAAGCAA------------------CAACAATTAGCACAACCTTCACATCATCAACATTACTATCAGCATCAACAACAACAGCAACAATAAATGCAACAT------TA----------------------------AA---------TGCAGCAGCAGCCGCACCACCAC---------------------------CAGTGCCGTCTGTTTGTGTGCGTGTGCCCCACA-------GTCCC |
| droAna3 |
scaffold_13266:4685454-4685690 - |
|
AA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTATAAC-----ATCCA----------------ACT----------------------ATATACGTCATC-AACTACAACA--ACAGCAGTTCCAAC---------------------AAC------AA------TATCTACAACA------------------------------ATACAACCAACAA---CTACAACAACAACAACAATTAGCACAACCTTCACATCATCAACATTACTATCAGCATCAACAACAACAGCAACAATAAATGCAACAT------CC----------------------------GC------------AGCAGCGGCCGCACCACCACCGC---------------------CGTCTGTGCCGTCTGTTTGTGTGCGTGTGCCCCACA-------GTCCC |
| droBip1 |
scf7180000395751:493368-493602 - |
|
AA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAATAAC-----ATCCA----------------ACT----------------------AA--ACGTCATC-AACTACAACA--ACAGCAGTACCAAC---------------------AAC------AA------CAGCAACAACA------------------------------ATACAAGCAACAG---CTACAACAACAACAACAATTAGCACAACCTTCACATCATCAACATTACTATCAGCATCAACAACAACAGCAACAATAAATGCAACAT------CC----------------------------GC------------AGCAGCGGCCGCACCACCACCAC---------------------AGTCTGTGCCGTCTGTTTGTGTGCGTGTGCCCCACA-------GTCCT |
| dp5 |
3:7222511-7222879 + |
|
GAGA---GCTCTCTGGGG-CGGACACTCGGGGTGA--------ATGTGT---------------------C---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C----------------------G----CTGGCAG-AGCTTCCACCGTACAGCCACCCACACACACATTACCTATACAC---GC-CCACGT------------AAA-------CTCGTTTAATCGTCTAATGTTATG----TGTGAATGTAGCC--AATAAAACAATAAC-----CA-GA----------------A--------------------------CTACATCATC-GACTACAACA--AC---------------------------------------------------AGCAACAACA------------------------------GTACAAGCAACAA---------CAACAGCAACAATAAGCACTACTTTCACATCATCAACATTATTATCAGCATCTACTACAACAGCAACAATAAACGCTGGATCTGCCCCG----------------------------TA------------TGCAGCAGCAGGGGCAG------GGGCAGGCGCCTCCATTGCAC---CAGTGCCGTCTGTTTGTGTTCGTGTGCCCCATA-------GTCCC |
| droPer2 |
scaffold_2:7422281-7422504 + |
|
A--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAATAAC-----CA-GA----------------A--------------------------CTACATCATC-GACT-TGACA--ACAG---------------------------------------------------CAACAACA------------------------------GTACAAGCAACAA---------CAACAGCAACAATAAGCACTACTTTCACATCATCAACATTATTATCAGCATCTACTACAACAGCAACAATAAACGCTGGATCTGCCCCG----------------------------TA------------TGCAGCAGCAGGGGCAG------GGGCAGGCGCCTCCATTGCAC---CAGTGCCGTCTGTTTGTGTTCGTGTGCCCCATA-------GTCCC |
| droWil2 |
scf2_1100000004590:1606752-1607016 + |
dwi_1792 |
GATG---AGGAACAGGCG-GAGGTTGGCGTAATAG--------ATCCAT---------------------CATTTATG-----C-----GAG--------------------------CTC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------G---------------------------------------------------------------------------------------------------------------CTATGTTCAACC--------------A----------------CCGATGCATCACCACATGCAAGATGATAGTGGGCTGC-AACTGCAACA--TCAACATCATCAAC---AACATC------AAC---ATC------AT------CAACAACATCA------------------------------GCTTCAACAA------------------CATCAGCATCAACAACATCAGCATCAACAACATCAGCATCAACAACATCAGCATCAACAACAACAACAACAGCTG------CA----------------------------AC------------TGCAGCAGCAGCAACAACAA---------------------------CAGC----------------------------------------- |
| droVir3 |
scaffold_12875:16130407-16130629 - |
|
TA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCATATCGTATAATG----TATG--TGT--------------------------------------------------------------------------------------------------AGCCAATACAACAACATCAACAACAATACAATCAAC------AACTAA---AACAACAACA------------------------------AC---AACAA------------AAACTACAACAATTAGCACGACTTTCACATCATCAACATTATTATCAGCATCAGCATCAACAACAACAACA---------G------CA----------------------------ACCGCTTTGAATCA---AGCGGCC---ATTGCAC---------------------------CAGTGCCGTCTGTTTGTGTTCGTGTGCCCCACA-------GTCCC |
| droMoj3 |
scaffold_6540:27478683-27478883 - |
|
ATGA---ATG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CC-----CAGCA----------------A-C----------------------AGCAACAGCAGC-AGCTGCAGCA--CCAGCAACACCAAC---------------------AGC------AGCAGC---AGCAACAGCA------------------------------GC---AACAA------------CAACAGCAACAACAACAGCAGCATCAACAACAACAACAGACACAGCAGCAACAGCAACAGCAACAACAACAGCTGCGACAG------AC----------------------------AC------------CGCCGC--CCACA--ACCAC---------------------------AA-C------A-----------ATGGCCA-T--GTCCACAG--CC |
| droGri2 |
scaffold_14830:2581255-2581487 - |
|
CC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AACAACAT---------------------------------------------------------CAGCAAC-AACACCAACA--ACAGCAACACCAAC---------------------AGCAGCAGCAACAAC---AACAACAACA------------------------------ACACCTACAACAA------------CAGCAGCAACAACAGCAACACCAACAACAACAGCAACACTATCAACAACATGCACTACAACAACAACACCAACAACTG------CCTGGCAAAATCTCATATCGCGGCATTTTCAC------------CACCAC--------------------------------------AGGCAACAACAACAGCAACAACATGAGCGCCGCACA-------G---C |