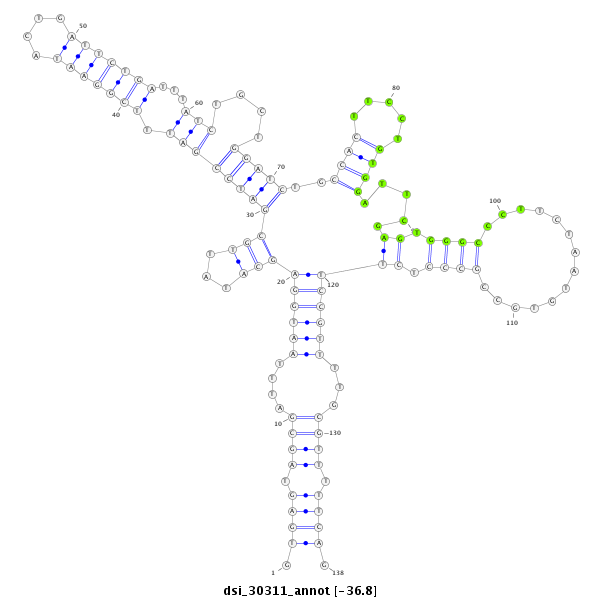
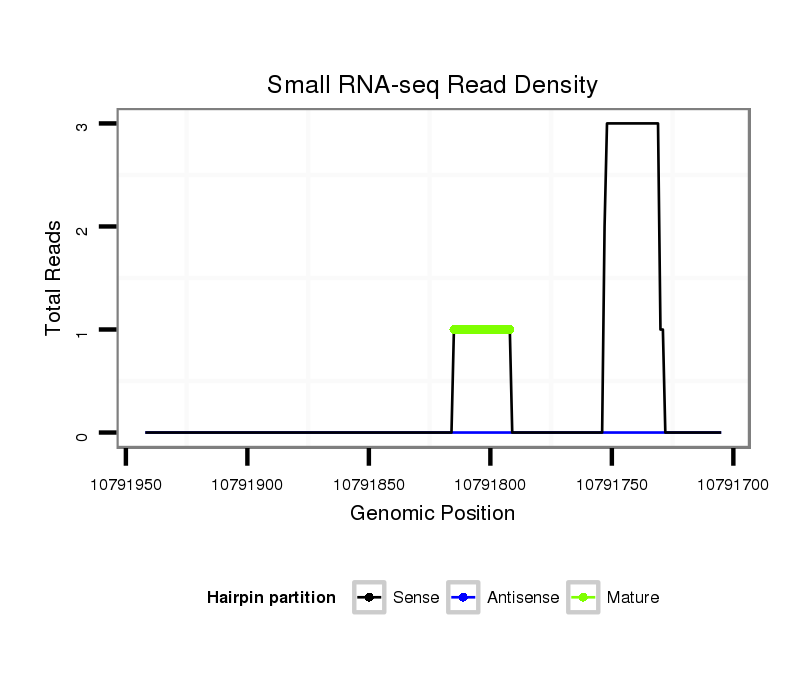
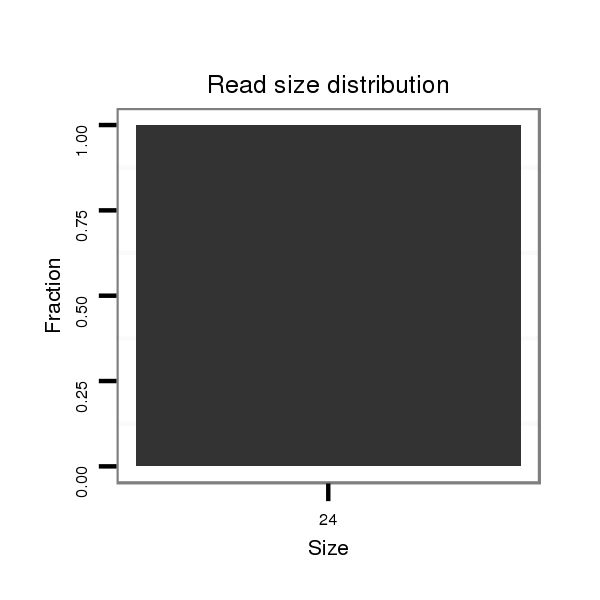
ID:dsi_30311 |
Coordinate:3r:10791755-10791892 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
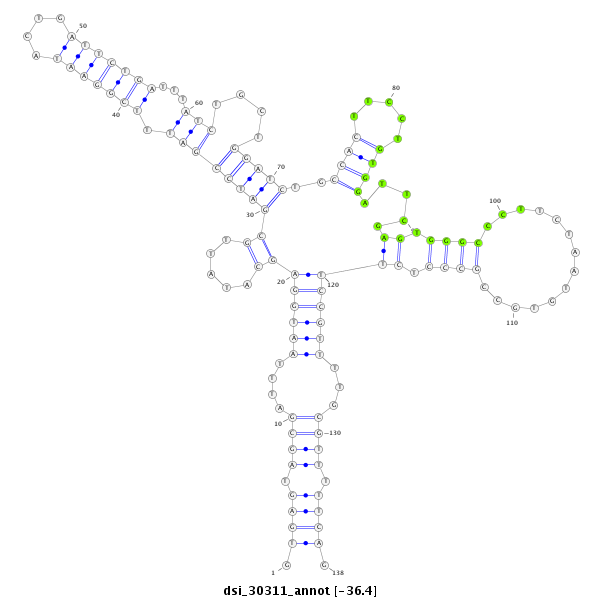
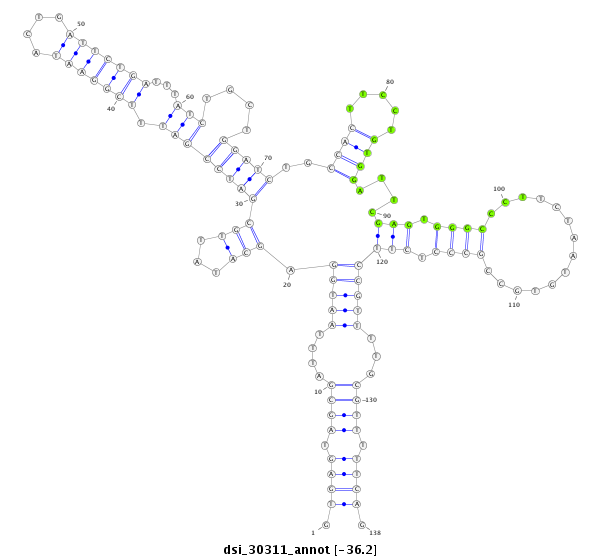
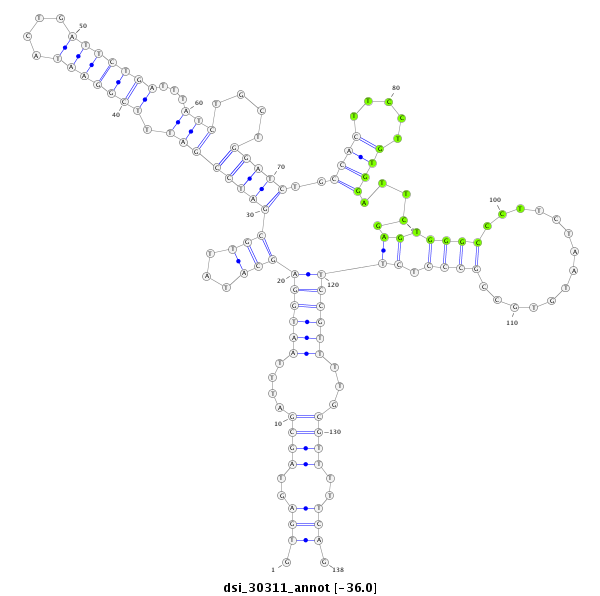
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -36.4 | -36.2 | -36.0 |
 |
 |
 |
exon [3r_10791573_10791754_-]; CDS [3r_10791573_10791754_-]; CDS [3r_10791893_10792131_-]; exon [3r_10791893_10792131_-]; intron [3r_10791755_10791892_-]
No Repeatable elements found
| ##################################################------------------------------------------------------------------------------------------------------------------------------------------################################################## CATTAGGAAGTCCGCGGGGCAGCTTCAATTCCCTCGCCGGCTATCCGCAAGTGAGTAGCGATTTAATGGAGCATATTGCGATCCGATTTCGGAATACTGATTCTGATTTATCTGCTGGATCTGCCACTTCCTGTGGATTCGAGTGGGCCCTTCTAATGTGCCGCCCTCTTCCGTTTTGCGTTTTTCAGCACGCGATTGAACTGCAGACGATGAACATGTACCGCCAGCGATCTGCGAA **************************************************.((((.((((....(((((((((...)))((((((((.(((((((....)))))))...)))....)))))..((((.....)))).....((.((((..............)))).))))))))...)))).)))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
M025 embryo |
SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
GSM343915 embryo |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................ACGCGATTGAACTGCAGACGATG.......................... | 23 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................TTCCTGTGGATTCGAGTGGGCCCT....................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................CGCGATTGAACTGCAGACGATGAA........................ | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................TGCGATCCGAGTTCGGAAT............................................................................................................................................... | 19 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................TGCGATCCGAGTTCG................................................................................................................................................... | 15 | 1 | 19 | 0.26 | 5 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TTATCTGCTGGAATTGCCACCT............................................................................................................. | 22 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................TGTAACGCCAGCGATCTACA.. | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................TATCTGCTGGAATTGCCAC............................................................................................................... | 19 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................GCCCTCTATCCTCAAGTGA........................................................................................................................................................................................ | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................AACTGCTGGATCTGTCA................................................................................................................ | 17 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................GAACAAGTAGCGCGAGCGA........ | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................................................................................................................................................................CGATGAACATGAACCC............... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
|
GTAATCCTTCAGGCGCCCCGTCGAAGTTAAGGGAGCGGCCGATAGGCGTTCACTCATCGCTAAATTACCTCGTATAACGCTAGGCTAAAGCCTTATGACTAAGACTAAATAGACGACCTAGACGGTGAAGGACACCTAAGCTCACCCGGGAAGATTACACGGCGGGAGAAGGCAAAACGCAAAAAGTCGTGCGCTAACTTGACGTCTGCTACTTGTACATGGCGGTCGCTAGACGCTT
**************************************************.((((.((((....(((((((((...)))((((((((.(((((((....)))))))...)))....)))))..((((.....)))).....((.((((..............)))).))))))))...)))).)))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
M024 male body |
|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................CTGACTTGACGTCTCCT............................ | 17 | 2 | 20 | 0.20 | 4 | 4 | 0 |
| .......................................................................................................................AGACGGTTAAGTACACCAAAG.................................................................................................. | 21 | 3 | 6 | 0.17 | 1 | 1 | 0 |
| .................CCGTAGAAGTTTAGGGAG........................................................................................................................................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 1 |
| .......................CAGTTAAGGGAGAGGCC...................................................................................................................................................................................................... | 17 | 2 | 9 | 0.11 | 1 | 1 | 0 |
| ...............................GGAGCGGCCTATAGGG............................................................................................................................................................................................... | 16 | 2 | 20 | 0.10 | 2 | 2 | 0 |
| .................CCTTAGAAGTTAAGGGAGT.......................................................................................................................................................................................................... | 19 | 3 | 16 | 0.06 | 1 | 1 | 0 |
Generated: 04/24/2015 at 10:49 AM