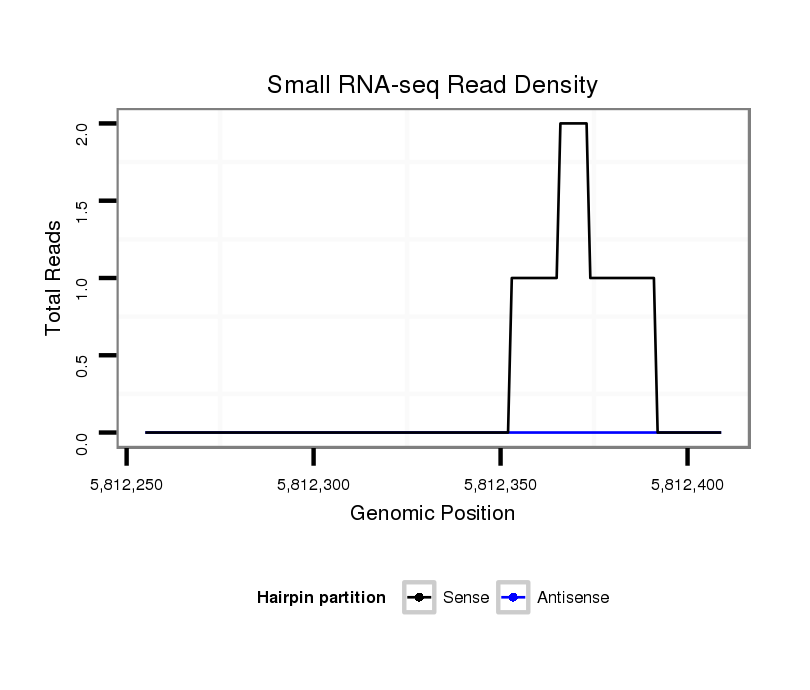
ID:dsi_30261 |
Coordinate:2r:5812305-5812359 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
exon [2r_5811572_5812304_+]; CDS [2r_5811572_5812304_+]; CDS [2r_5812360_5812451_+]; exon [2r_5812360_5812451_+]; intron [2r_5812305_5812359_+]
No Repeatable elements found
| ##################################################-------------------------------------------------------################################################## GGCTACCGAAATGGCCAGCAAACTGTACGAGGAAAAGGACGAATTACTTGGTGAGTATGTGAAATAAATTATGCCCTACTTAGTGAGTAATGCCAGTTTTACCAGTTCGAGTCGTGGAGGTTCTTGGCATAGATGTGTGCCTAGAGCTATACAAA **************************************************........(((((((.......((..(((((...)))))..))..)))))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR902009 testis |
SRR553488 RT_0-2 hours eggs |
M024 male body |
M053 female body |
SRR553485 Chicharo_3 day-old ovaries |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................TGGAGGTCCTTGGCATAGATGA................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................TTACCAGTTCGAGTCGTGGAG.................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................TCGTGGAGGTTCTTGGCATAGATGTG.................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................TCCCCCGTTCGAGTCGTGGA..................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................GAAGAAAATGACGAATTGCTT.......................................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................GTTCGAGTCGCGGACG................................... | 16 | 2 | 20 | 0.10 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................GTTCGATTCCTGGAGGTAC................................ | 19 | 3 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................TCAGTTCGTGTCGTGGATG................................... | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................ACGAGGAAAAGGAAGACT............................................................................................................... | 18 | 2 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................GCAAGTCGTGGAGGTT................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................GTTCGAGTCTCGGAGG................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CCGATGGCTTTACCGGTCGTTTGACATGCTCCTTTTCCTGCTTAATGAACCACTCATACACTTTATTTAATACGGGATGAATCACTCATTACGGTCAAAATGGTCAAGCTCAGCACCTCCAAGAACCGTATCTACACACGGATCTCGATATGTTT
**************************************************........(((((((.......((..(((((...)))))..))..)))))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
M024 male body |
|---|---|---|---|---|---|---|---|---|---|
| ..............GGTCGTTTCAGATGCCCCTTTT....................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .....GGCTTTGACGGTCGTTTAACAT................................................................................................................................ | 22 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 |
| .................................................................................................................................AACTACCCACGGATCTC......... | 17 | 2 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 |
| ............................................................................ATGATTGACTCACTACGGT............................................................ | 19 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................TCCAAGAAGCGTATCTG..................... | 17 | 2 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 |
| ................TCGTTTGACAGGCCCGTTT........................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 12:05 AM