ID:dsi_3008 |
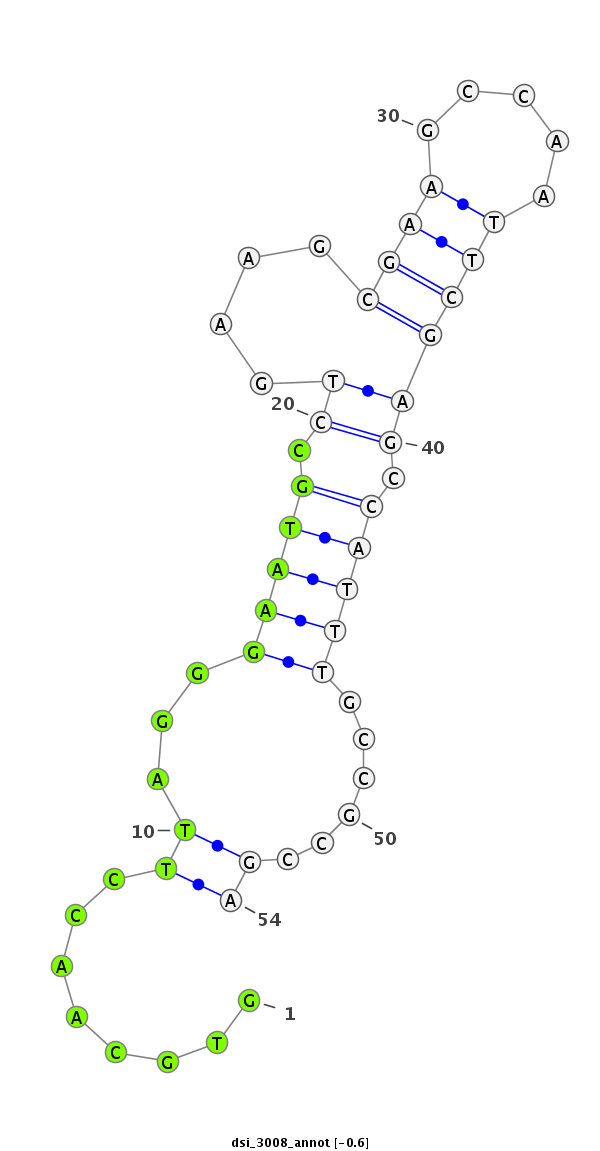
Coordinate:3l:11673021-11673074 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
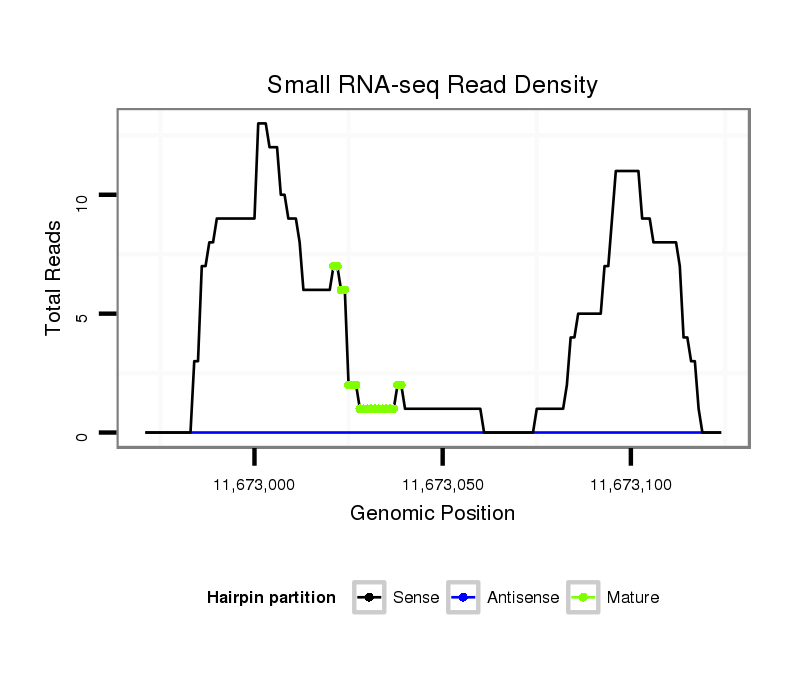
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
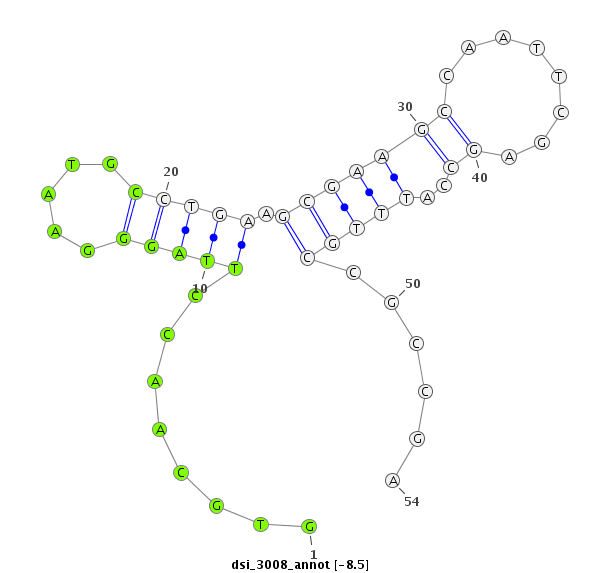
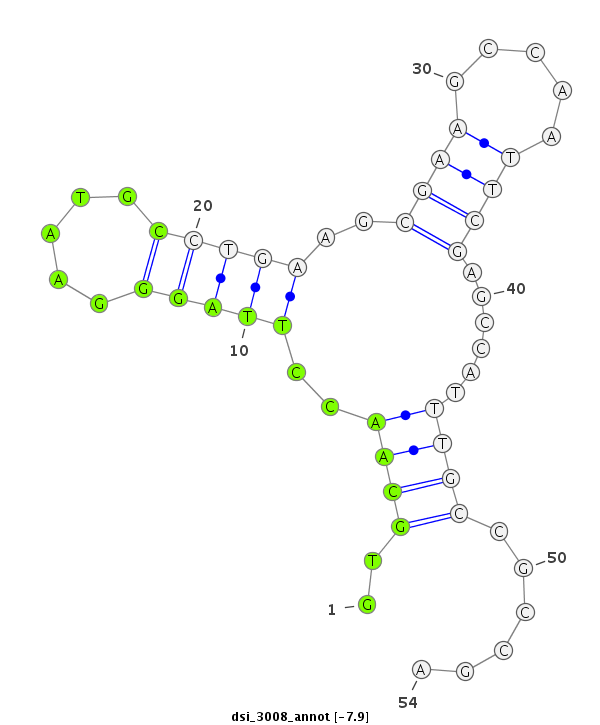
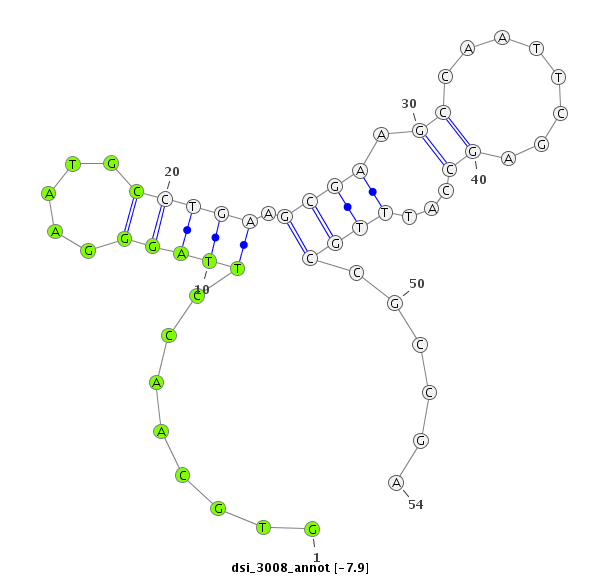
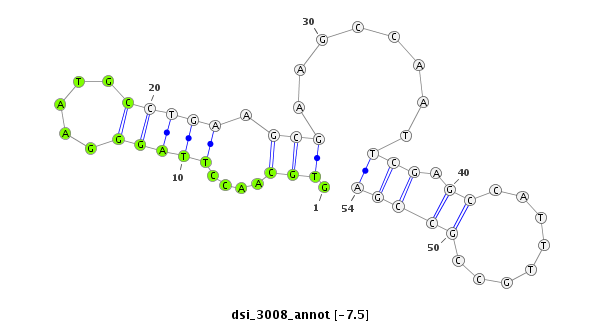
| -8.5 | -7.9 | -7.9 | -7.5 |
 |
 |
 |
 |
exon [3l_11672476_11673460_+]; CDS [3l_11672476_11673273_+]
No Repeatable elements found
| ########################################################################################################################################################## GACAAGGCGCAGGGTATATGTTTGCTGAAGGACGCCATGGCCATGATCTCGTGCAACCTTAGGGAATGCCTGAAGCGAAGCCAATTCGAGCCATTTGCCGCCGAGAACACCAAGTCCGGCTTCTGTTCCTGTGACGATCTGCAGCGCGTTAGTG **************************************************........((...(((((.((....((((.....)))))).)))))......))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
GSM343915 embryo |
SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR553485 Chicharo_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................TTCCTGTGACGATCTGCAGCGC....... | 22 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................CTGTTCCTGTGACGATCTGCA........... | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................GTCCGGCTTCTGTTCCTGT...................... | 19 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............ATATGTTTGCTGAAGGACGCC...................................................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................GACGCCATGGCCATGATCTCGTGC.................................................................................................... | 24 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............ATATGTTTGCTGAAGGACGCCATGGC................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................ATGGCCATGATCTCGTGC.................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................AGTCCGGCTTCTGTTCCT........................ | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............ATATGTTTGCTGAAGGAC......................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................GAACACCAAGTCCGGCTTCTGC............................ | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................GACGCCATGGCCATGATCTCGTGCAAC................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............GTATATGTTTGCTGAAGGACGCC...................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............GTATATGTTTGCTGAAGGACGCCAT.................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................GTGACGATCTGCAGCGCG...... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............GTATATGTTTGCTGAAGGAC......................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................GTTCCTGTGACGATCTGCAGC......... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................GTTCCTGTGACGATCTGC............ | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTGCAACCTTAGGGAATGC..................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................TCTCAGGCAACCCTAGGGAAT....................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................CACCAAGTCCGGCTTCTGTTCCC........................ | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................ATGTTTGCTGAAGGACGCCATGGCC................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................GCCATGGCCATGATCTCGTGC.................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................GAACACCAAGTCCGGCTTCT.............................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................GTTCCTGTGACGATCTGCA........... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............ATATGTTTGCTGAGGGACGCCATGGC................................................................................................................. | 26 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................GCCTGAAGCGAAGCCAATTCGAG................................................................ | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................CCGGCTTCTGTTCCTGTGAC................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................GACGCCATGGCCATGATCTCGT...................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................GTTTGCTGAAGGACGCCATGGCC................................................................................................................ | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................GACGATCTGTAGCGGGT..... | 17 | 2 | 9 | 0.33 | 3 | 0 | 0 | 2 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................GACGATCTGTAGCGGGTC.... | 18 | 3 | 20 | 0.30 | 6 | 0 | 0 | 2 | 4 | 0 | 0 | 0 |
| ..................................................................................................................................GCGACGATCTGTAGCG........ | 16 | 2 | 20 | 0.25 | 5 | 0 | 0 | 3 | 1 | 1 | 0 | 0 |
| .....................................TTGCCATGAGCTCGTGAAAC................................................................................................. | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................GACGATCTGTAGCGGG...... | 16 | 2 | 20 | 0.15 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................ACGATCTGTAGCGGGT..... | 16 | 2 | 20 | 0.15 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 |
| ..............................................................................AGCCAATTCGAGCGA............................................................. | 15 | 1 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
CTGTTCCGCGTCCCATATACAAACGACTTCCTGCGGTACCGGTACTAGAGCACGTTGGAATCCCTTACGGACTTCGCTTCGGTTAAGCTCGGTAAACGGCGGCTCTTGTGGTTCAGGCCGAAGACAAGGACACTGCTAGACGTCGCGCAATCAC
**************************************************........((...(((((.((....((((.....)))))).)))))......))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902009 testis |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................TTCAGGCCGAAGACTACGAAA...................... | 21 | 3 | 3 | 1.00 | 3 | 3 | 0 | 0 |
| .................................GGGTACCGTTACTAGAGCAGGT................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 |
| ..............................................................................................................GTTCAGGCCGAAGACTACTA........................ | 20 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 |
| ............................................................................CTTCCGTTAAGCTCG............................................................... | 15 | 1 | 16 | 0.06 | 1 | 0 | 0 | 1 |
Generated: 04/24/2015 at 10:34 AM