




ID:dsi_30076 |
Coordinate:2r:15708169-15708318 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -38.5 | -38.3 | -38.3 |
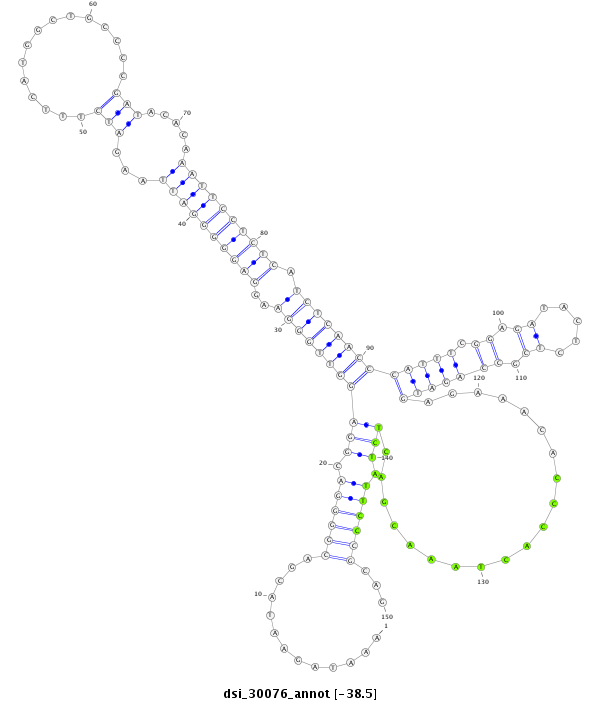 |
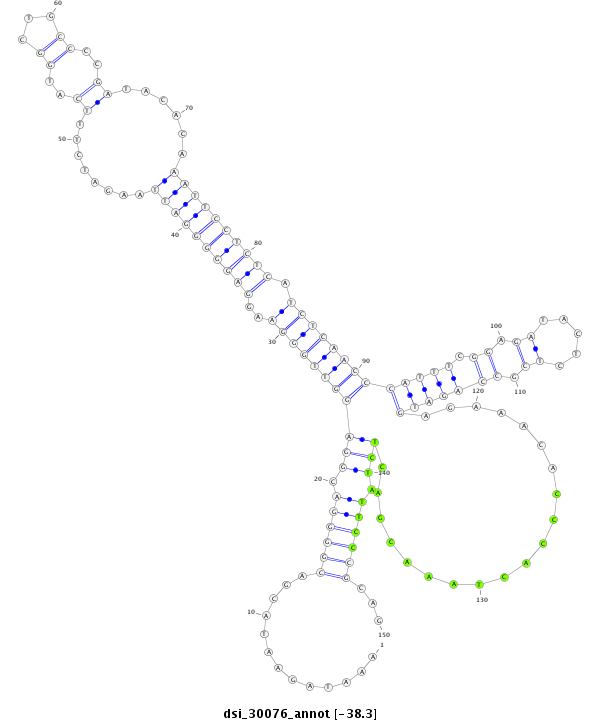 |
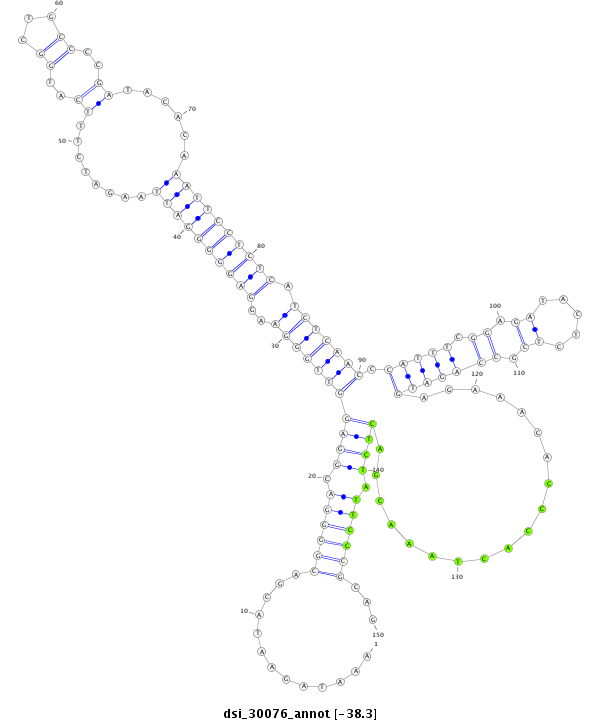 |
exon [2r_15708319_15708862_+]; five_prime_UTR [2r_15708319_15708373_+]; intron [2r_15701932_15708318_+]
No Repeatable elements found
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TTTTCCCCAACGTTTCATATTTTCCCCAAAGAAAATTGTACAAATTCTATAAATAGAATACGACGGGGACGGAGGTTGGGAAGGAGGGGGATTAAGATCTTTCATGGCTGCCCCGATACACAAATTCCTCTCATCTCAACCCATTTCGGAGATACTCTCGCCAGATGAGAAACACCCACTAAACGACTCTATTCCCGCAGTACGATAACGACGATGACGACGATCTGTGGGATTAACTCGGCCTGGAACT **************************************************.............((((((.(((((((((((..((((((((((...(((...............))).....)))))))))).))))))).(((((.((.((.....)).)))))))...................)))).))))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
SRR553487 NRT_0-2 hours eggs |
GSM343915 embryo |
SRR553488 RT_0-2 hours eggs |
M025 embryo |
SRR553486 Makindu_3 day-old ovaries |
M024 male body |
SRR902009 testis |
SRR618934 dsim w501 ovaries |
SRR553485 Chicharo_3 day-old ovaries |
M053 female body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................CCCACTAAACGACTCTATTCC....................................................... | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................ACCATCTGTGGGATTA............... | 16 | 1 | 3 | 1.33 | 4 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................GACGACGATCTGTGGGATT................ | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................TAAATAGAAAACGACGGGGACGGAGGT.............................................................................................................................................................................. | 27 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................GACGATCTGTGGGATGAACC............ | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................GACGACGATCTGTGGGAT................. | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................GATGACGACGATCTGTGGGATT................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................AGGAAGGATGGGGATTAAG.......................................................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................GATGACGACGATCTGTGGGAT................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................GATGACGACGATCTGTGGGATTA............... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................GCTACCATCTGTGGGATTA............... | 19 | 3 | 19 | 0.74 | 14 | 0 | 8 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................CTACCATCTGTGGGATTA............... | 18 | 2 | 3 | 0.67 | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................ACCATCTGTGGGATTAT.............. | 17 | 2 | 14 | 0.64 | 9 | 0 | 6 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................CTACCATCTGTGGGATT................ | 17 | 2 | 11 | 0.55 | 6 | 0 | 4 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................AATAGAATAGGACGGG....................................................................................................................................................................................... | 16 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................GATTAACTGGACCTGGAAC. | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................GAAGGATGGGGATTAAGAA........................................................................................................................................................ | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................AACGACGATCACGACGATCA........................ | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................ACCATCTGTGGGATT................ | 15 | 1 | 12 | 0.33 | 4 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................TACCATCTGTGGGATTA............... | 17 | 2 | 10 | 0.30 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................ACGGGGACGGTGGTGGGGA......................................................................................................................................................................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................GACGGGGTCGGTGGTGGGGA......................................................................................................................................................................... | 20 | 3 | 12 | 0.17 | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................AAAACGACGATCACGACGATCA........................ | 22 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| TTTTCCGAAACGTTTTATATTTT................................................................................................................................................................................................................................... | 23 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TGACGGGGACGGTGGTGGGGA......................................................................................................................................................................... | 21 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................AATACGACGGGGATGTAGC............................................................................................................................................................................... | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................................................................CTACCATCTGTGGGAT................. | 16 | 2 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................ACCGACGATCACGACGATCA........................ | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................CGGGGACGGTGGTGGGGA......................................................................................................................................................................... | 18 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................CTGTGGGATGAACCAGGCC....... | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................................................................................................GACGCTCTGTGGGATT................ | 16 | 1 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................................................................CTACCATCTGTGGGATTAT.............. | 19 | 3 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........AACATATCATACTTTCCCC............................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................................GAAGGATGGGGATTAAGTA........................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................ACGATATCGACGATCACGT.............................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
|
AAAAGGGGTTGCAAAGTATAAAAGGGGTTTCTTTTAACATGTTTAAGATATTTATCTTATGCTGCCCCTGCCTCCAACCCTTCCTCCCCCTAATTCTAGAAAGTACCGACGGGGCTATGTGTTTAAGGAGAGTAGAGTTGGGTAAAGCCTCTATGAGAGCGGTCTACTCTTTGTGGGTGATTTGCTGAGATAAGGGCGTCATGCTATTGCTGCTACTGCTGCTAGACACCCTAATTGAGCCGGACCTTGA
**************************************************.............((((((.(((((((((((..((((((((((...(((...............))).....)))))))))).))))))).(((((.((.((.....)).)))))))...................)))).))))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
M023 head |
M053 female body |
SRR618934 dsim w501 ovaries |
M025 embryo |
SRR553485 Chicharo_3 day-old ovaries |
SRR902009 testis |
GSM343915 embryo |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR1275487 Male larvae |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................CGGGGATATGGGTTTAAG........................................................................................................................... | 18 | 2 | 3 | 70.00 | 210 | 117 | 42 | 35 | 7 | 7 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................CGGGGATATGGGTTTAAGC.......................................................................................................................... | 19 | 3 | 12 | 17.58 | 211 | 136 | 29 | 40 | 0 | 4 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................CGGGGATATGTGTTTAAGCTG........................................................................................................................ | 21 | 3 | 2 | 3.50 | 7 | 6 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................CGGGGATATGCGTTTAAG........................................................................................................................... | 18 | 2 | 4 | 1.50 | 6 | 2 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................CGGGGATATGTGTTTAAG........................................................................................................................... | 18 | 1 | 2 | 1.50 | 3 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................CTGCTACTGCTGCTAGACACC.................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................CGGGGATATGTGTTTAAGC.......................................................................................................................... | 19 | 2 | 2 | 1.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................GCGGGGATATGGGTTTAAG........................................................................................................................... | 19 | 3 | 16 | 0.94 | 15 | 3 | 1 | 10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................CGGGGATATGGGTTTAA............................................................................................................................ | 17 | 2 | 12 | 0.42 | 5 | 0 | 0 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................................GGGGATATGGGTTTAAG........................................................................................................................... | 17 | 2 | 18 | 0.17 | 3 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................ATGTGTTTACGGAGAGGA.................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................TAAGAAGAGTAGGGCTGGGT........................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................GGCGGGGATATGGGTTTAAG........................................................................................................................... | 20 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................CGGGGATATGAGTTTAAGC.......................................................................................................................... | 19 | 3 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................TGAGATCGGGGCGTCA................................................. | 16 | 2 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .............................................................................................................CGGGGATATGTGTTAAAGC.......................................................................................................................... | 19 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................AAGTTTCGACGGGGCTA..................................................................................................................................... | 17 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................GGGGATATGGGTTTAA............................................................................................................................ | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................AAGGAGTGTAGAGATGCGT........................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................TTCTAGCTAGTACCGA............................................................................................................................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 2r:15708119-15708368 + | dsi_30076 | TTTTCC--CCAACGTTTCATATTTT-CC--CCAAAGAAAAT--------------------------TGTACAAATTCTATAAATAGAA------TACGACGGGGACG-----------GAGGTTGGGAAGG---A---------------GGGGGAT-TAAGATCTTTCATGGCTGC----------CCCGATACACAAAT------TCCTCTCATC-TCAA-------CCC-------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTTC--GGAGAT-ACTCT-CGC-------------------C-------AG--ATGAGAAA--CACCCA-CTA-AACGA----------------------CTCT--A-------------T------TCCCGCAGTA---CGATA-----A-------CGAC---GATGACGAC---GATCTGTGGGATT---------------AACTCGGCCTGGAACT |
| droSec2 | scaffold_1:12570672-12570914 + | TTTTCA---AAACGTTTCATATTTT-CC--CCAAAGAAAAT--------------------------TGTACAAATTCTATAAATAGAA------TACG---GGGACG--------------GTTGGGGAGG---A---------------GGGGGAT-TAAGATCTTTCATGGCTGC----------CACGATACACAAAT------TCCTCTCATC-TCAA-------CCC-------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTTC--GGAGAT-ACTCT-CGC-------------------C-------AG--ATGAGAAA--CACCTA-CTA-AACGA----------------------CTCT--A-------------T------TCCCGCAGAA---CGATA-----A-------CGAC---GATGACGAC---GATCTGTGGGATT---------------AACTCGGCCTGGAACT | |
| dm3 | chr2R:15057925-15058172 + | TTTTCC---CAACGTTTCATATTTTTCC--CCAAAGAAAAT--------------------------TGTACAAATTCTATAAATAGAA------TACGACAGAGACG--------------TTTGGGCAGG---AG--------------GGGGGAT-TAAGATCTTTCATGGCTGC----------CCCGATACACAAAT------TCCTCTCATC-TCAA-------CCC-------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTAC--GGAGAT-ACTCT-CGC-------------------C-------AG--ATGAGAAA--CACCCA-CTA-AAAGA----------------------CTCT--A-------------T------TCCCGCAGAA---CGATA-----A-------TGAC---GATGACGAC---GATCTGTGGGATT---------------AACTCGGCCTGGAACT | |
| droEre2 | scaffold_4845:9257989-9258242 + | TTTTCC---CAACGTTTCATATTTT-CCCCCAAAAGAAAAT--------------------------TGTACAAACTCTATAAATAGAA------TACGACGGGGACG--------------GTTGGCTAGC---A---------------GGGGGAT-TAAGATCTTTCATGGCTGC----------CCCGATACACAAAT------TGCTCTCATC-TCAA-CCTCATCCC-------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTTC--GGAGAT-ACTCT-CGC-------------------C-------AG--ATGAGAAA--CACCCA-CTA-AAAGA----------------------CTCT--A-------------T------TCCCGCAGAA---CGATA-----A-------CGAC---GATGACGAC---GATCTGTGGGATT---------------AACGCGGCCTGGAACT | |
| droYak3 | 2R:14121240-14121509 - | TTTTTC--CCAACGTTTCATATTTT-CCCCCAAAAGAAAAT--------------------------TGTACAAATTCTATAAATAGAA------TGCGACGGCGACG--------------GTTGGTTAGGAGGA---G-------GAGGAGGGGAT-TAAGATCTTTCATGGCTGC----------CCCGATACACAAAT------TCCTCTCATC-ACAC-A----ACCC-------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTTC--GGAGAT-GCTCTCCGC-------------------C-------AGAGAAGAGAAA--CACCCA-CTA-AAAGA----------------------CTCTATA-------------TTCCCATGCCCGCAGAA---CGATA-----A-------CGAC---GATGACGAC---GAACTGTGGGATT---------------AACACCGCCTGGAACT | |
| droEug1 | scf7180000409663:247880-248143 - | TTTTTT---CACCGTTTCATATTTT-CC--CAAAAGAAAAT--------------------------TGTACAAGTTCTATAATTAGAA------TGTGCTGAGCATGAAAGGGGAGTGGAGATTGAG----AAGCG--------------GGGGACT-TTAGATCTTTCATGGCTGC----------CCCGATACACAAAT------TTCTCTCATC-CCAA-A-----ACC-------------------------------------------------------------------------------------------------------------------------------------------------------------------ATCTC--AGCGATTTCTCT-CGC-------------------C-------CG--ATGAGAAA--CACCCA-CTATAATGA----------------------CTAT--A-------------T------TCCCGCAGAA--CCGACA-----A-------CGAC---GATGACGAC---GATCTGTGGGATT---------------AACTCGCCCTGGAACT | |
| droBia1 | scf7180000302292:1283410-1283663 - | TTTTTC---CAACGTTTCTTATTTT-CC--CAAAAGAAAAT--------------------------TGTACAAGTTCTTTAAATAGAA------TGCGATGGGGATT-----------------G-GGAGCTAGA---------------GGGGGAT-TTAGATCTTTCATGGCTGC----------CCCGATACACAAAT------TCCTCTCATC-TTAA-------CCC-------------------------------------------------------------------------------------------------------------------------------------------------------------------GTCTCGCGGAGAT-GCTCT-CGC-------------------TGCGGTGATG--ATGAGAAA--CACCCA-CTA-AAAGA----------------------CTCT--A-------------T------TCCCGCAGAA---CGACA-----A-------CGAC---GATGACGAC---GATCTGCGGGATT---------------AACGCGCCCTGGAACT | |
| droTak1 | scf7180000415386:176113-176356 + | TTTTTT---CAAC-TTTCATATTTT-CC--CAAAAGAAAAT--------------------------TGTACAAGTTCTTAAAATAGAA------TGCGATGAGGATG------------------GGTAGCAAGA---------------GGGGGAT-TTAGATCTTTAATGGCTGC----------CCCGATACACAAAT------TCCTCTCATC-TCAA-------CCC-------------------------------------------------------------------------------------------------------------------------------------------------------------------ATCTC--GGAGAT-GCTCT-CGC-------------------C-------CG--CTGAGAAA--CACCCA-CTA-AAAGA----------------------CTCT--A-------------T------TCCCGCAGAA---CGACA-----A-------CGAT---GATGCCGAC---GATCTGTGGGATT---------------AACTCGCCCTGGAACT | |
| droEle1 | scf7180000491009:79972-80233 - | TTTTTT---CAACGTTTCATATTTTCCC--CAAAAGAAAAT--------------------------TGTACAAGTTCTATAATTAGAT------AGCGATGAGGATG--------------TTGGGACAGC---AGGGGAAACGGGGGGAAGGGAAG-GGAGATCTTTCATGGCTGC----------CCCGATACACAAAA------TCCACTCGCC-TCAA-------CCC-------------------------------------------------------------------------------------------------------------------------------------------------------------------ATCGC--GGAGAT-TCTCT-CGC-------------------C-------AA--GTGAGAAA--CACCCA-CTA-AAAGA----------------------CTCT--A-------------T------TCCCGCAGAA---CGACC-----A-------CGAC---GATGACGAC---GATCTGTGGGATT---------------AACTCGCCCTGGAACT | |
| droRho1 | scf7180000780106:196540-196776 - | TTTTTC---CAACGTTTCATATTTT-CC--CAAAAGAAA----------------------------------AGTTCTATAATTAGAT------AGCGATGAGAATG------------------GGGAGCAACA---------------GGGGGAG-GCAGATCTTTCATGGCTGC----------CCCGATACACAAAA------TCCTCTCGCC-TCAT-------CCC-------------------------------------------------------------------------------------------------------------------------------------------------------------------ATCTC--GGAGAT-TCACT-TGC-------------------C-------AA--GTGAGAAA--CACCCA-ATA-AAAGA----------------------CTCT--A-------------T------TCCCGCAGAA---CGACA-----A-------CGAC---GATGACGAC---GATCTGTGGGATT---------------AACTCACCCTGGAACT | |
| droFic1 | scf7180000454044:870347-870589 + | TTTTTC---CAACGTTTCATATTTT-CC--CAAAAGAAAAT--------------------------TGTACAAGTTCTATAATTAGAA------TGCGTTGAGGATG------------------GGGCTT---T---------------GGGGGAT-GTAGATCTTTAATGGCTGC----------CTCGATACACAAAA------TCCTCTCACCCTCTA--------AA-------------------------------------------------------------------------------------------------------------------------------------------------------------------ATCTC--GGAGAA-TCGTT-CGC-------------------C-------GA--ATGAGAAG--CACCCA-CTA-TATGA-----------------A----CTAT--A-------------T------TCCCGCAGAA---CGACA-----A-------TGAT---GATGACGAC---GATCTGTGGGATT---------------AACTCGCCCTGGAACT | |
| droKik1 | scf7180000302682:864643-864903 + | -----------ACGTTTCATATTTTTCC--CAAAAGAAAATT-------GTTTTTGT----------TGTACAAATTCTATAATTAGAA------CGAGTTGAG----------------------------------------------------------GATCTTTCATGGCTGTGCCCCGCCGACCCAATACACAAAA------TAGTCT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CT-CGC-------------------C-------AAA-ATGAGAGAGCCA-CCA-CTA-CCAACATCAGCCAGCACCTTTCACTAATTCG-TATCCCATCCATCCATTTGTA-TCCCACAGAA---CTACT--ACGACGAACCACGAC---AATGCCGAC---AATCTGTGGGATT---------------AACGCTCGCTGGAACT | |
| droAna3 | scaffold_13266:11500931-11501155 - | CTTTCCAACGAACGTCTCATAGGTT-CC--CCAACGAAAATC-------------------------CGAACGGATTCTATAAATAGGG------C-----GGGGACT-----------G-----GGGCAGGCGGC--------------CGGGGGAG-GGAG-CCGTTCATGGCTGC----------CCCGATACAGAAAA------TTGTCTCGC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T-------CT--CTCAGATA--CACGAATCTA-AAAGA----------------------TACT----------------T------TCCCGCAGAC---CGACT--CCGC-------CGAC---GATGACGACGCTGATCCGGGGGATT---------------ATCGCGCGCCGGAACT | |
| droBip1 | scf7180000396547:967058-967275 + | TTTTTC--TGAACGTCCTATAGCTT-CC--CAAATGAAAATC-------------------------TGAACAGATTCTATAAATAGAG------CG-----GGGACT-----------G-----GGGCAGGAACT---------------TGGGGGT-TGGGGGCT-------CTGC----------CCCGATACAGAAAA------TCATCTCGC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T-------CT--CTGAGATA--CACGAAACTA-TAAGA----------------------TACT----------------T------TCCCACAGAC---CGACACTCCGA-------CGAC---GATGACGACGTTGATCCGTGGGATT---------------ACCTCGCGCCGGAACT | |
| dp5 | 3:9891076-9891367 + | TTTTT------------CATATTTT-CATTTTCATGAAAAT--------------------------TGTTCGAGTTCTATAAATAGACACGCCCAGCGGTGGGGCTG-----------C-----GGGTAGGAAGG---------------GAGGGATGGGAGGTGTTTCATGGC-----------------ATACAGAAACCATGGCATCTCTCATC-TGT-----------------------------------------------------------------------------------------------------------------TGCTCTCT----------------CTCTCTCTATCTCTGTC------------TGTGGCACATACC--ACGAGT-ACTCT-GGCTCTCGTATAATACAACTAT-----------------------------TTC-TCTGT----------------------CTGA--T-------------C------TTCGACAGAACACCGACT-----A-------CGAC---GATGAT------GAACCAACGCCTGGACCTCGGCTGGAACTCGTCCGTCTGCTGCT | |
| droPer2 | scaffold_4:5209115-5209404 + | TTTTT------------CATATTTT-CATTTTCATGAAAAT--------------------------TGTTCGAGTTCTATAAATAGACACGCCCAGCGGTGGGGCTG-----------C-----GGGTAGGAAGG---------------GAGGGATGGGAGGTCTTTCATGGC-----------------ATACAGAAACCAAGACATCTCTCATC-TGT-----------------------------------------------------------------------------------------------------------------TGCTCTCT------------------CTCTCTATCTCTGTC------------TGTGGCACATACC--ACGAGT-ACTCT-GGCTCTCGTATAATACAACTAT-----------------------------TTG-TCTGT----------------------CTGA--T-------------C------TTCGGCAGAACACCGACT-----A-------CGAC---GATGAT------GAACCAACGCCTGGACCTCGGCTGGAACTCGACCGTCTGCTGCT | |
| droWil2 | scf2_1100000004822:1754210-1754473 + | TTTTTT---TTTT-------------T---------AAAAATGTTCATAATTTTTGCCAGAGAAAATT-------TGCTATAAATAGAC------TTGGGGGG---------------------------------------------------------------------------------------------------------------------------------------------------------------CTACACATACACATAGAAGGAAATATAT---ATACAAAGAGATATATGTATATATATC------------GGAAGATCTCT------------------CTCTTTTTCCTTGTTTCTCTCTCAGTTT-------GTCTA--ATAA-T-TGTCT-CTC-------------------TTTTTGACTG--TTTTGCAG--CATTCA-ACA-AAAAA----------------------AAAT--A----------------------------------TATC-----A-------AAATAATGATGATTCC---TTGCTGGAATTTG---------------ATTTCTATCTGTTAAT | |
| droVir3 | scaffold_13324:2701658-2701912 + | TTTTT------------CATATTTT-C----AAACGAAAAAT----------TTAAA----------AAAAAAGAATCTATAAATAGGC------AC-----GG----------------------------------------------------------GTCTTTTCATGGCTGC----------CCA----TAGAGAT------CTCAGAGATC-TGAG-A-----GCTCCCAGAGCCAGTGGTGCTACGTTG--TTG------------------------------------------------TAGATAC---CGACTATA----------TGTTTCGAT----------ATCG-----AATTCGAAGT------------TGCAACTCATTCC--GTGA-----------------------------------------------------------------TTGT----------------------TCCG-AT-------------TTGCAG-----GCTTAACTGGGATT-----T-------TAAT---GATGCTGAC---CGCCCCGTGGATC---------------GTCGCCATGCGCTGCT | |
| droMoj3 | scaffold_6496:14092555-14092830 - | dmo_1146 | TTTTT------------CATATTTT-CTAATGAAAAAAAAAT-------------------------GTTCAAAGTTCTATAAATAGAC------ATGGCTATACACA-----------------------------------------------------------------------------------------GAGATCGCAGCAG-----------AGCA-----GATCCCCGAGCCAGTGGCGCT-TGTTG--TTGCAATACACATACACACAAACACACACAC---ACACACTTAGATACACCTAAATATACTGTTATATATGGGCCA-ACTGTTTCGAATTACTAACTAATCC-----------------------------------------------------------------------------------------------------------------AA----------------------TTCG-AA-------------TTTCAGACTTTG--TTG---CCATT-----A-------TGAT---GCTGCTGTC---TGATCGCTGGACC---------------TGCACCATGCTCTGTT |
| droGri2 | scaffold_15112:4770516-4770788 - | TTTTTT-----------TATATATC-CCAATGAACAAATATT----------T--------------TTTTAAATATCTATAAATAGAC------GT-----GGGACT-----------TCTGTTG-----GCCGA---------------GAGAGAT-C--------------------------------------------------------------------------CCCGAGCCATACACAGA-TTTTGACTGCTTACACAGACACACACAACCACACACACAACATACACACACTTACACGTAAATGT---GGCCTATATGTGCCAAACTGTTTCGAT----------ATCA-----------------------------------TTTTC--GAAGTT-ATTC---AC-------------------T-------GG--GT---------------TTA-ACTGT----------------------TTCC-AC-------------TTTCAG-TCCG---GCA---CGATT-----A-------TAAT---GATGATGTT---GT---------------------------------AATGATGTT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/18/2015 at 12:11 PM