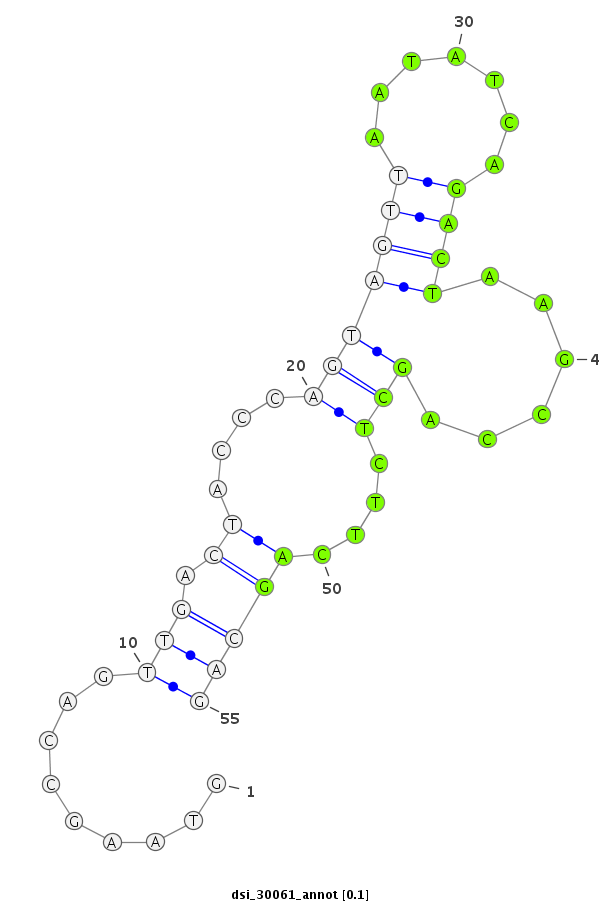
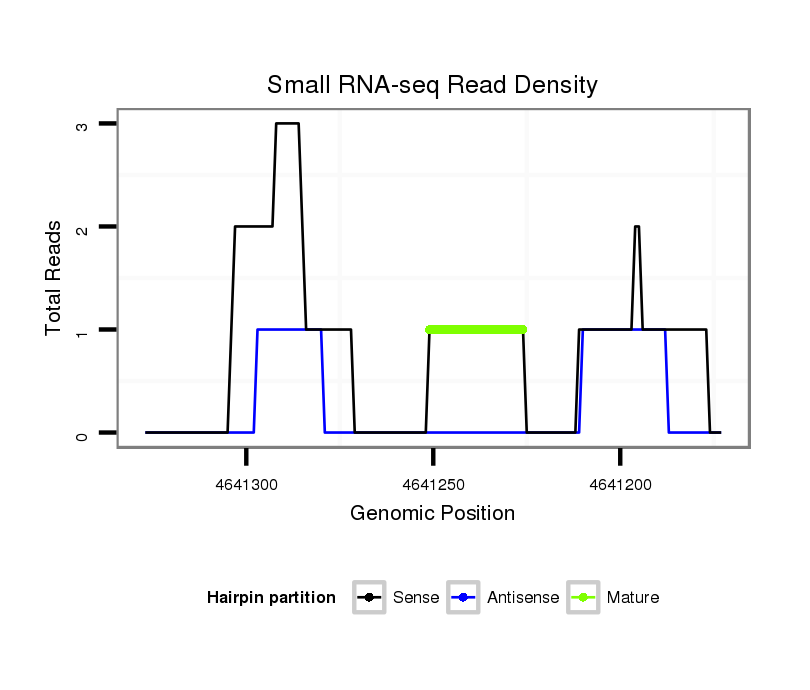
ID:dsi_30061 |
Coordinate:3r:4641223-4641277 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -7.9 | -7.1 | -7.0 | -6.9 |
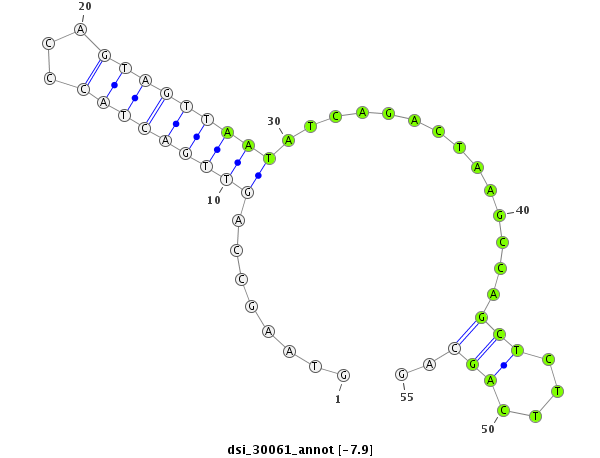 |
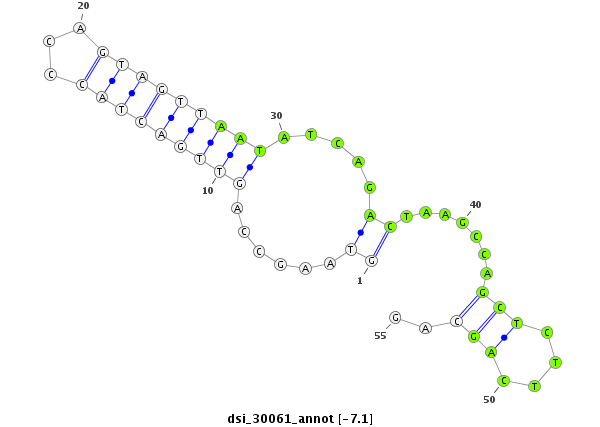 |
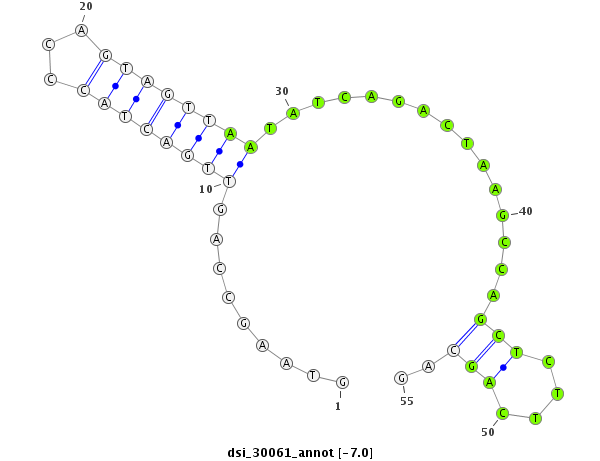 |
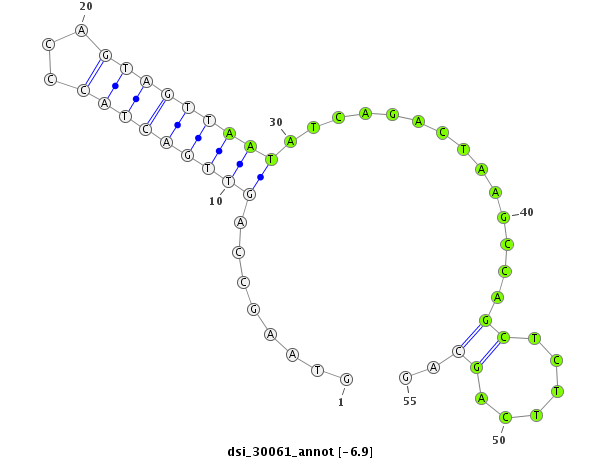 |
exon [3r_4641014_4641222_-]; CDS [3r_4641014_4641222_-]; CDS [3r_4641278_4641388_-]; exon [3r_4641278_4641388_-]; intron [3r_4641223_4641277_-]
No Repeatable elements found
| ##################################################-------------------------------------------------------################################################## CAGCACCCATCACGCTCATCAACCAGCGACCTGTGGCCGAGGATGTGCACGTAAGCCAGTTGACTACCCAGTAGTTAATATCAGACTAAGCCAGCTCTTCAGCAGGAATACCCCCAGACAAACAGCCTGAAGAAGAACACCAGCACCTTGAGCAG **************************************************.........(((.((....(((((((.......))))......)))....)))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902009 testis |
GSM343915 embryo |
M025 embryo |
SRR553485 Chicharo_3 day-old ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
M023 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................GAAGAACACCAGCACCTTGA.... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................GCCGAGGATGTGCACGTAAGC................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................AGCGACCTGTGGCCGAGG................................................................................................................. | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................AGCGAACTGTGGCCGAGGATT.............................................................................................................. | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................CAGCGACCTGTGGCCGAGGA................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................GACAAACAGCCTGAAGA...................... | 17 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................AATATCAGACTAAGCCAGCTCTTCAG..................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................AAGCGAACTGTGGCCGAGGATTT............................................................................................................. | 23 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................AGCGAACTGTGGCCGAGGTT............................................................................................................... | 20 | 2 | 12 | 0.25 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................AGCGAACTGTGGCCGAGGTTTT............................................................................................................. | 22 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................AGCGAACTGTGGCCGAGGTTT.............................................................................................................. | 21 | 3 | 16 | 0.13 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................CGAACTGTGGCCGAGGTTA.............................................................................................................. | 19 | 3 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................AGCGAACTGTGGCCGAGGTTA.............................................................................................................. | 21 | 3 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................GAACAAGAACACTAGCCCC........ | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GTCGTGGGTAGTGCGAGTAGTTGGTCGCTGGACACCGGCTCCTACACGTGCATTCGGTCAACTGATGGGTCATCAATTATAGTCTGATTCGGTCGAGAAGTCGTCCTTATGGGGGTCTGTTTGTCGGACTTCTTCTTGTGGTCGTGGAACTCGTC
**************************************************.........(((.((....(((((((.......))))......)))....)))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
M023 head |
SRR553487 NRT_0-2 hours eggs |
SRR902009 testis |
M024 male body |
SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
M053 female body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................TATGGGGATCTGTTGCTCGG............................ | 20 | 3 | 3 | 1.00 | 3 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................TCATCAATTATAGTATGTTTCAGT.............................................................. | 24 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................GACACCGGCTCCTACACG........................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................TGTTTGTCGGACTTCTTCTTGTG............... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................TCGGTTTTCTTCTTTTGGTCGT.......... | 22 | 3 | 11 | 0.73 | 8 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................GGGGGTCAGTTTGACGGA........................... | 18 | 2 | 7 | 0.29 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ............................................................................................................ATGGGGATCTGTTACTCGG............................ | 19 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................ACCGGATCCTACTCGTGC........................................................................................................ | 18 | 2 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................TCGGACTTCTTCTGG................. | 15 | 1 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................TTGGTTGCTGGGCACCG...................................................................................................................... | 17 | 2 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................AGTTTGTCGCTGCACAC........................................................................................................................ | 17 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................GGGTCAGTTTGTCGG............................ | 15 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................ACGCTCGACACCGGCTCC................................................................................................................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 10:17 AM