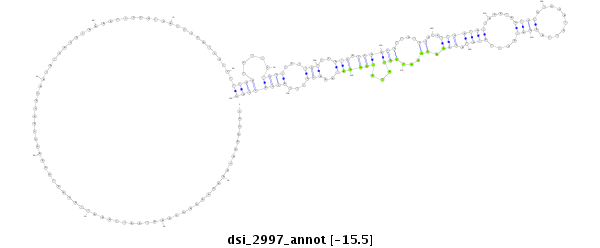
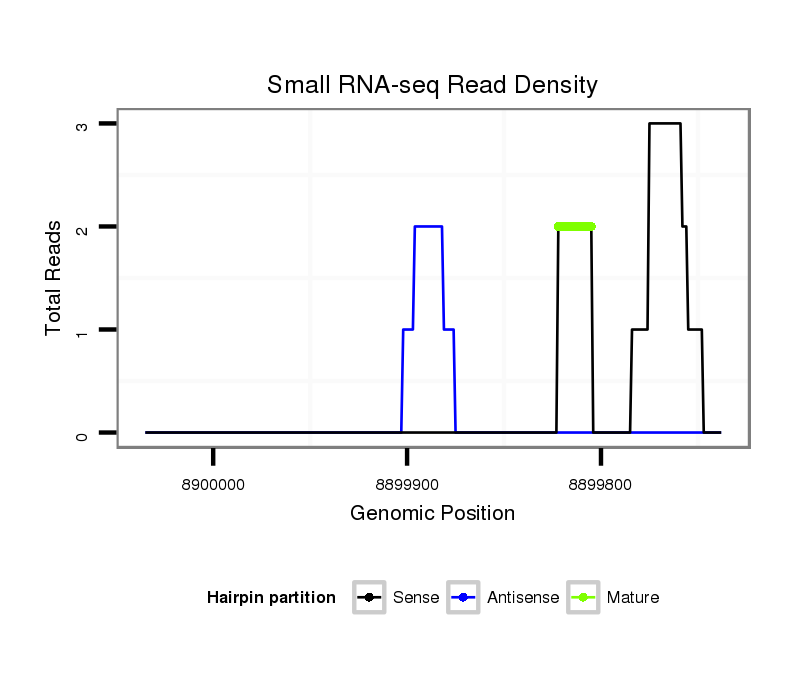
| droSim2 |
3l:8899738-8900035 - |
dsi_2997 |
TTTAA---------------ATCGGAACACACAACAATAA------------------T--ATA--------CAA--GC-AAACACCACCAA-A--TATCGG-----------AAAAG----AAAG-------------------------AAGAGCAAATAATTAAATTACAAT--AGAA-ACCAAGAGAA----------------------------------TATCACGC------AAAA----T---------CC--CATC--------ATCCCAT-CGTCTT-TGGTCTTCTGGTTCTTGTTCATCACCAACGT----T---------------------------------ATC-GATAT-TGATATTAAATAG-----TCTGTGGATAT--TAAGATACTTATATACAAACGCATCG--TATCTGGGATAAACGTTTACCACCAGGAATATTAATGATACG--------------------------------------------------------CA------------------ACGG-----------------------CTG---------TAACTGTAAACTGTGTCCCCTCTCA--- |
| droSec2 |
scaffold_0:1385786-1386083 - |
|
TTTAA---------------ATCGGAACACACAACAATAA------------------T--ATA--------CAA--GC-AAACACCACCAA-A--TATCGG-----------AAAAG----AAAG-------------------------AAGAGCAAATAATTAAATTACAAT--AGAA-ACCAAGAGAA----------------------------------TATCACGC------AAAA----T---------CC--CATC--------ATCCCAT-CGTCTT-TGGTCTTCTGGTTCTTGTTCATCACCAACGT----T---------------------------------ATC-GATAT-TGATATTAAATAG-----TCTGTGGATAT--TAAGATACTTATATACAAACGCATCG--TATCTGGGATAAACGTTTACCACAAGGAATATTAATGATACG--------------------------------------------------------CA------------------ACGG-----------------------CTG---------TAACTGTAAATTGTGTCCCCTCTCA--- |
| dm3 |
chr3L:9123081-9123384 - |
|
TTTAA---------------ATCGGAACACACAACAATAA------------------TATATA--------CAA--GT-AAACACCACTAA-A--TATCGG-----------AAAAG----AAAG-------------------------AAGAGCAAATAATTAAATTACAAA--AGAA-ACCAAGAGAA----------------------------------TATCAAGC------TAAA----T---------CC--CATCATCCCATCATCCCAT-CGTTTT-TGGTCTTCTGGTTCTTGTTCATCACCGACGT----T---------------------------------ATT-GATAT-TGATATTGAATAC---------TGGATAT--TAAGATACATATATACAAACGCATCG--TATCTGGGATAAACGTTCATCACAAGGAATATTAATGATACG--------------------------------------------------------CA------------------AAGG-----------------------CTG---------TAACTGTAAACTGTGTCCCCTCTCA--- |
| droEre2 |
scaffold_4784:4921446-4921731 + |
|
TTTAA---------------ATCGGAACACACAACAATAA------------------A--ATA--------CAT--GC-AAACACCACCAA-A--TATCGG-----------AAAAG----AA--------------------TAGAAA-AAGAGCAAATTATAAAATTACGAT--AGAA-ACCAAGAGAA----------------------------------TATTAACC------GAAA----A---------CC--CGTCA-------ATCCCAT-CGTCTT-TGGTCTTCTGGTTCTTCTTCATCACCAACGTCCGTTAAG---------------------A-ACCACGCATC-GTTAT-TG------AATAA-----TCTGTGGATAT--TGAGATACATATATA----CGCATCG--CATCTAGGATA----------------------AATGATACG--------------------------------------------------------CA------------------ACGA-----------------------CTG---------TAACTGTAAACTGTGTCCCCTCTCA--- |
| droYak3 |
3L:2794233-2794549 + |
|
TATAA---------------ATCGGAACACACAACAATAA------------------A--ATA--------CAA--GC-AAACACCACCAA-A--TATCGG-----------AAAAG----AAAA---------TAA------TAGAAAAGAGAGCAAATAATAAAATTACGAA--AGAA-CCCAAGAGAA----------------------------------TATCACGC------TAAA----A---------CC--CGTC--------ATCCCAT-CGTATT-TGGTCTTCTGGTTCTTGTTCATCACCAACGTCCGTTAAG---------------------A-ACCACGCATC-GATAT-TG------AATAA-----TCTGTGGATAT--TGAGATACATATACACGTACGTATCG--TATCTAGGATAAACGCTCACCAGCACGGATATTAATGATACG--------------------------------------------------------CA------------------ACGG-----------------------ATG---------TAACTGTAAACTGTGTCCCCTCTCA--- |
| droEug1 |
scf7180000409467:328677-328943 - |
|
TATAA---------------ATCGGAACAC--AACAAAAA---------------------ATA--------CAA--GT-AAACACCACCAAAA--TATTGA-----------AAAAT----T-AGAAGAAAAAA-----------TTACAACT----------------------------ACCAAGAGAA----------------------------------TATCAACC------ATAA----A---------CT--CGCC--------ATCCCAT-CGTCTT-TGGTCTTCTTGTTCTTGTTA------AACGTCCGTTGGA-----------------------ACCACGCATC-GTGAT-TG------AATAATA-----G----ATCT--GGAGAT------------------TGTATATCTATGATAAACGTATA---CGAGAGATATAGATGATACG--------------------------------------------------------TA------------------ACTGCT-----------------GCA------A---CTATAACTGTAAACGGTGTCC--TCTCA--- |
| droBia1 |
scf7180000300910:2082102-2082389 + |
|
TATAA---------------ATCGGAACAC--AACAATAA---------------------ATA--------CAA--GC-AAACACCACCAA-G--TATCGA-----------AAA-T----AGAG--AAATAA-A----------TTTAAAAA----------------------------AACAAGAAAT----------------------------------TATCCATC------CTAA----A---------CT--CGTC--------GCCCCAA-CGTCTTTTGGTCTTCTTGTTCTCGTTC------AACGTCCGTTAAA---------------------C-ACCACGCATC-AAGAT-TG------AGTAACATCATCTGTGGATATCCCGAGATATATAGATACA----------ATATCTGTGACAAGCGAGTGC---CAGAGATATAGATGATAAG--------------------------------------------------------GA------------------AAG-CC-------------ACTTGCC------A---CTGTAACTGTAAACGGAGTCCC--CTCA--- |
| droTak1 |
scf7180000415356:62603-62893 + |
|
TATAA---------------ATCGGAACAC--AACAATAA---------------------ATA--------CAA--GC-AAACACCACCAA-A--TATCGA-----------AAAG-----AAA-------------------TAGAAAGAAAAAC----------------------------AAGA--------------------------------------ATCCATC------TTAA----A---------CT--TGTC--------GTCCCAT-CGTCTTTTGGTCTTCTTGTTCTTGTTC----TCAACGTCCGTTAAA---------------------A-ACCACGCATC-AGGAT-TG------AATAATA-----T----ATAT--CGAGATATATACAT-------------ATATCTATGACAAGCGTATAC---CAGAGATATAGATGATAAGCAGCAGCAGCAGCAGC--------------------------------------------------------------AGCGTTCGG-GAAAAGCACTTGCA------A---CTGTAACTGTAAACGGTGTCCC--CTCA--- |
| droEle1 |
scf7180000491193:3364889-3365146 - |
|
ATA-ATTCCATGTCAAT-AAATCGGAACAC--AACAATAAATACACCACC---------------------------AA-A----------A-A--TATCAA-------------AAG----AAAA-------AAT----------TATATATA----------------------------ATCAAGAGAA----------------------------------TATCAACA------T-AAACCAACCAA-CGCACC--CGTC--------GTCCCAT-CGTCTT-TGGTCTTCTTGTTC------------AACGTTCGTTAAA-----------------------ATCCCGCATC-GAGAT-AG------AGCTATA--------------------------------------------CATCTGTAATATG---------CAAGATATATAGATGATACG--------------------------------------------------------TA------------------ATAGCTTTCTG-GA------ATTGCCGCTG---------CAACTGTAAACTGTGTCCC----CA--- |
| droRho1 |
scf7180000777034:198909-199191 - |
|
ATA-ATTCCATCTTTAT-AAATCGGAACAC--AACAATAA---------------------ATA--------CAA--TA-AAACACCACCAATA--TATCTA-----------AAAGA----GAAA-------------------------AATTTTAAAA------AATATA-TATAAAACATTAAAAGAA----------------------------------TATCAACA------T-AAACAAACCAGCCGCACTCGTGTC--------GTCCCAT-CGTCTT-TTGTCTTCTTATTC------------AACGTTCGTTAAA-----------------------ATCACGCATC-GAGAT-TG------AGTTTT--------------------------------------------ATATCTATGATATG---------CAAGATATATAGATGATACG--------------------------------------------------------TA------------------ATAGCTTTTGG-AA------ACGGCAACTG---------CAACTGTAAACTGTGTCCC----CA--- |
| droFic1 |
scf7180000454065:2544989-2545267 + |
|
TATAA---------------ATCGGAACAC--AACAACAA---------------------ATA--------CAA--ACCAAACACCACCAAAA--TATCGA-----------AAAAA----AAAG-------------------------AAA---AAATAAA-AAAATACA-T--CAAA-ACCAAGAGAA----------------------------------TATCAAAG------TAAA----ATCAACCACA----GTTC--------GTCCCAT-CGTCTT-TGGTCATCTTGTTCTTGTTC------AGCGTCCGTTTAA-----------------------GCCACGCATCCGAGAT-TG------AATAACA-----TATTTATAT--AG--------------AA--------GATATATATGATATG---------CGA-AGATATAGCTGATACG--------------------------------------------------------TA------------------ATAGCATCCGAAGA------A------TTG---------CAACTGTTAACTGTCTCTC----CA--- |
| droKik1 |
scf7180000302486:1631714-1632009 - |
|
A-TAA---------------ATCGGAACAC--AACAATAA---------------------AC----CAA--CAAGGAA-AAACACCACCAA-A--TATCGA-----------GGAGCATCAAAAA------------ATCGTT--TAAATAGAA---------------GCAAT--AAAA-GCTAGAAAAACAACTAAAGAT------ATAGTATCTCTACCGTGTTTCATCC------AAAA----TCCAACCGCA-T--CGTA--------GTCCCATTCGTC-T-TGGTCATCTAGTTCTTGTTCTTGTTCAGCAT------------------------------------------------TG------AGTAA----------------------------ATATACA-GCGTATTG--TATTT---ATATGCCTGTGCCAT---------------CACG--------------------------------------------------------TAATCGGTAAATTCGGC---AT--------------------TGCA------ATTCCTACAACTGTAAACGGTATCTG----AA--- |
| droAna3 |
scaffold_13337:19524174-19524432 - |
|
ATA-ATTCCATCAAAAT-AAATCGGAACAC--AACAATAA---------------------ATA--------CAA--GC-AAACACCACCAA-A--TATCCG-----------AAAAA----AAAA------------TTCCTT--TAAAAAAA--------------------------------------------AACGAAAAAAAAAAG-------------TATCT----------ACT----A---------CT--CGTC--------G--CCAT-CGTTTT-TGGTCTTCTTGT----GTTC---------ATCCGTTTCAAAAAAAAAAAGAAATAAAAATAAAAAACGCATC-GAGAT-TG------AATAATA-----TTTA--------------------------------G--TATCTGGCAT----------------------------CACG--------------------------------------------------------CA------------------AC--------------------------TGCAA---TTATAACTGTGAATTTCAACGCGAGCGACAA |
| droBip1 |
scf7180000396360:718952-719193 - |
|
CATCA--------AAATAAAATCGGAACAC--AACAATAA---------------------ATA--------CAA--GTAAAACACCACCAAAATATATCTGAAATATATTCGAAAAG----AAA-------------TT-------------------------------CA-T--CAA--------AGAA----TAAACGA------AAAG-------------TATCT----------ACA----A---------CG--CGTC--------G--CCAT-CGA--T-CGGTCTTCTTGTT---GTTC------AA---CCGTTTAA--------CTGAAAG-AAAATA-AAAACGCATC-GAGATTTG------AATAAAA-----TTTA----------------------------------ATATCTGGCAT----------------------------CACG--------------------------------------------------------CA------------------AC--------------------------TGCAATACCAACGTCTGT---------GAACTTCAA--- |
| dp5 |
XR_group5:558137-558200 - |
|
AATAATTCGAAACG---AATATCGGGACAA--CACAAATA---------------------ATACAACAGTAAAC--GA-AAACACCACCAG-A-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droPer2 |
scaffold_67:175418-175481 + |
|
AATAATTCGAAACG---AATATCGGGACAA--CACAAATA---------------------ATACAACAATAAAC--GA-AAACACCACCAG-A-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droWil2 |
scf2_1100000004837:1474153-1474231 + |
|
AAAAA---------------AAAAAAATAA--AATA------------------------------------TAT--G-----TTCTAATAA-A--TAAAAA-----------AAAA--------A-------A-A--------TTTAAAAAAAAGCAAA--ATCGAAACACAAC--AAAA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droVir3 |
scaffold_13049:1591489-1591756 + |
|
TTCAA---------------ATCGGAACGC--AACATAAGA--------------------AAACAACAACAAAC--AA-GAACAACA--------CACTCG-----------A-------------------------------------ACC----------------------------ACCA----AT----------------------------------TAACAACATTAACCGTTA----ACC-GTCGCGCATCCGTC----------------AAACTT-TAACCGTCTTTTACGC------------------------------------------------------------AT-TG------AGTAA-----TAAGAGCACAA--AG---------------------------------------------------AAAAAATAGAAAAGAAAAAAAAACTAAAGCAGCAACGTAACTCCGTAACGTATCAAGAACGCGTCCCGTCTCCCGAATTGTAATTGTGTGTGTGCGA-----------------------ATG---------CAACAGCAACAAGAATCTCTCTTCA--- |
| droMoj3 |
scaffold_6654:2233713-2233775 - |
|
TTCAA---------------ATCGGAACGC--AACATAAGACAAAACAACAACAACAAC----------------------------------A--TACTCG-----------AAAAC----CA-----------------------------------------------------------CCA----A----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTA |
| droGri2 |
scaffold_15110:2677365-2677429 + |
|
TTGAA---------------ATCGGAACCC--CGCAAAACACAAGGA--------------AA-------------------ACA-------------------------------------------------------------TAAAACAA----------------------------AACAAGAAAA----------------------------------AACCACCA------A--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTA |