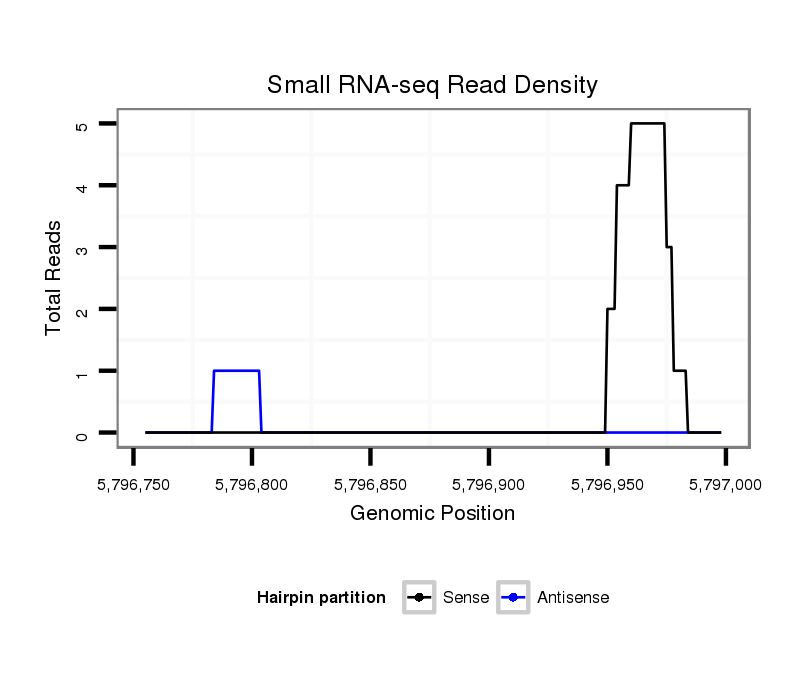
ID:dsi_29867 |
Coordinate:2r:5796805-5796948 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
exon [2r_5796612_5796804_+]; CDS [2r_5796612_5796804_+]; CDS [2r_5796949_5797544_+]; exon [2r_5796949_5797544_+]; intron [2r_5796805_5796948_+]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| ##################################################------------------------------------------------------------------------------------------------------------------------------------------------################################################## GCATTTGGGTCTCAAACGACTTCGAAATTGCACCCGAAGATACCCAGCTTGTAAGTTACCGTATAATTATTAATTTATTTATTAAAAGCAATCTTGAAATATAAAGTATTATAATTGATATGAGCACTATCGGAGCTCATAATTAACTCCTTATCATAAATTAGTGCGTAATGACTTATTACACTTTTGCTCAGTACAAAACGACTATGGAAGCTCTGCTGCAGACCTCTGAACCCAGTGCAAG **************************************************((((((((.(((((......((((((((..........((((..(((.((.....)).)))..)))).(((((((.((...)).)))))))..............)))))))))))))...))))))))...............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
M024 male body |
SRR553487 NRT_0-2 hours eggs |
SRR902009 testis |
M053 female body |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
GSM343915 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................AACGACTATGGAAGCTCTGCTGCA..................... | 24 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................ACAAAACGACTATGGAAGCTCTGCT........................ | 25 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................AAACGACTATGGAAGATCTGCC........................ | 22 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................TATGGAAGCTCTGCTGCAGACCTC............... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................TGACTATGGAAGCTCTGCTGCAGACCTC............... | 28 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................AAACGACAATGGAGGTTCTGC......................... | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................TATGAGCACTATCGA............................................................................................................... | 15 | 1 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................AAACCCAGCTTGAAAGTT........................................................................................................................................................................................... | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................ATATGAGCACTGTCGAA.............................................................................................................. | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................................................................................AGGAAACTCTGCTGAAGAC.................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CGTAAACCCAGAGTTTGCTGAAGCTTTAACGTGGGCTTCTATGGGTCGAACATTCAATGGCATATTAATAATTAAATAAATAATTTTCGTTAGAACTTTATATTTCATAATATTAACTATACTCGTGATAGCCTCGAGTATTAATTGAGGAATAGTATTTAATCACGCATTACTGAATAATGTGAAAACGAGTCATGTTTTGCTGATACCTTCGAGACGACGTCTGGAGACTTGGGTCACGTTC
**************************************************((((((((.(((((......((((((((..........((((..(((.((.....)).)))..)))).(((((((.((...)).)))))))..............)))))))))))))...))))))))...............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
SRR553486 Makindu_3 day-old ovaries |
GSM343915 embryo |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|
| .............................CGTGGGCTTCTATGGGTCGA................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................TCGTGTTTTGCTGATA.................................... | 16 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................................................TTTTTTGCTGATACCTAC............................... | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 |
| ....................................TTCTATTGGTCGAACAC............................................................................................................................................................................................... | 17 | 2 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 |
| ...........................................................GGATATTAATAATTAA......................................................................................................................................................................... | 16 | 1 | 15 | 0.07 | 1 | 0 | 0 | 0 | 1 |
Generated: 04/23/2015 at 11:40 PM