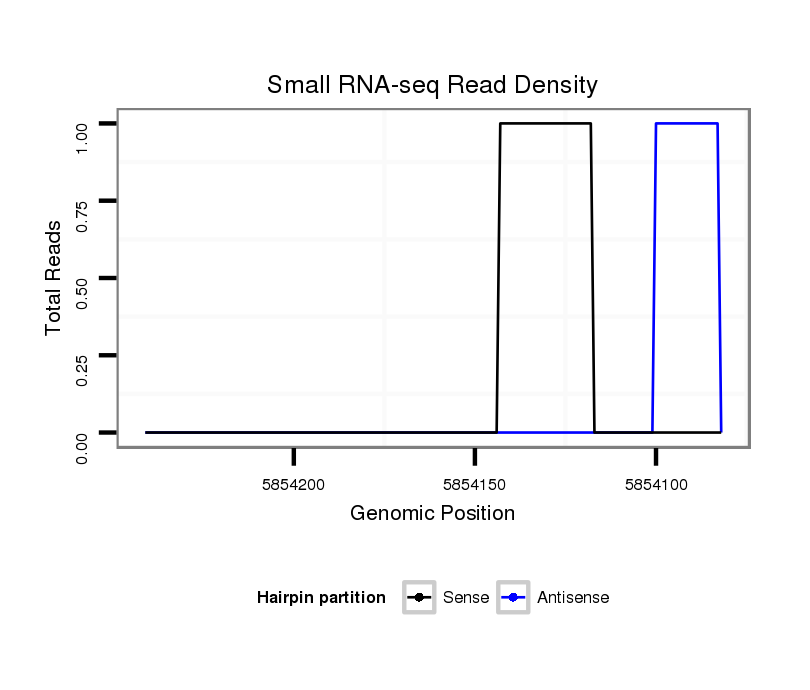
ID:dsi_2933 |
Coordinate:2r:5854132-5854191 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
exon [2r_5853942_5854131_-]; CDS [2r_5853942_5854131_-]; CDS [2r_5854192_5854337_-]; exon [2r_5854192_5854337_-]; intron [2r_5854132_5854191_-]
No Repeatable elements found
| ##################################################------------------------------------------------------------################################################## CCGCAGTGGCTGGTTCAGGTGGAGTTCTCTTCATATGGTCGCTCCGCAAGGTGAGTTGTATGTGAAGCCGGCTAACTAGATGCTTGACCTGTGACCCCTTTAACTGACAGTGCACCAATTCGGCAATCCTGTGGCTGGCACTGGTGTCCCTGCTGAGCCA **************************************************((.(((((........((.((.(((........))).)).))........))))).))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR902009 testis |
M024 male body |
|---|---|---|---|---|---|---|---|---|---|
| ............................................................TGTGAAGCTGGCTAACTAGATGCT............................................................................ | 24 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ...........................................................................................................CAGTGCACCAATTCGGCGATC................................ | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................TTTAACTGACAGTGCACCAATTCGGC.................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................TAACTGACAGTGCACCAATTCGGCGAT................................. | 27 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................ACTGAGAGTGCACCA........................................... | 15 | 1 | 7 | 0.86 | 6 | 0 | 6 | 0 | 0 |
| ...CAGTGGATGGATGAGGTGGAGTT...................................................................................................................................... | 23 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| ......................................................................................................ACTGAGAGTGCACCATTGCGG..................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 |
| ......................................................................................................ACTGAGAGTGCACCACT......................................... | 17 | 2 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 |
| ........................................................................................CTGTGACCTCTTTGACCGA..................................................... | 19 | 3 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................GTTGACTGAGAGTGCACC............................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................................GTGCACCACGTCGGCAAC................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| .............................................................................................................................................TGCTGTCACTGCTGAGC.. | 17 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
|
GGCGTCACCGACCAAGTCCACCTCAAGAGAAGTATACCAGCGAGGCGTTCCACTCAACATACACTTCGGCCGATTGATCTACGAACTGGACACTGGGGAAATTGACTGTCACGTGGTTAAGCCGTTAGGACACCGACCGTGACCACAGGGACGACTCGGT
**************************************************((.(((((........((.((.(((........))).)).))........))))).))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | O001 Testis |
GSM343915 embryo |
SRR618934 dsim w501 ovaries |
M024 male body |
M023 head |
|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................ACCACAGGGACGACTCGG. | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................CTGGCGAAATGGACTGTCA................................................. | 19 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................ACTGTGGAAATGGACTGT................................................... | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................TGAAATGGACACTGGAGAAA........................................................... | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................CTGGGCTGTCACGTGGTT.......................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 01:26 AM