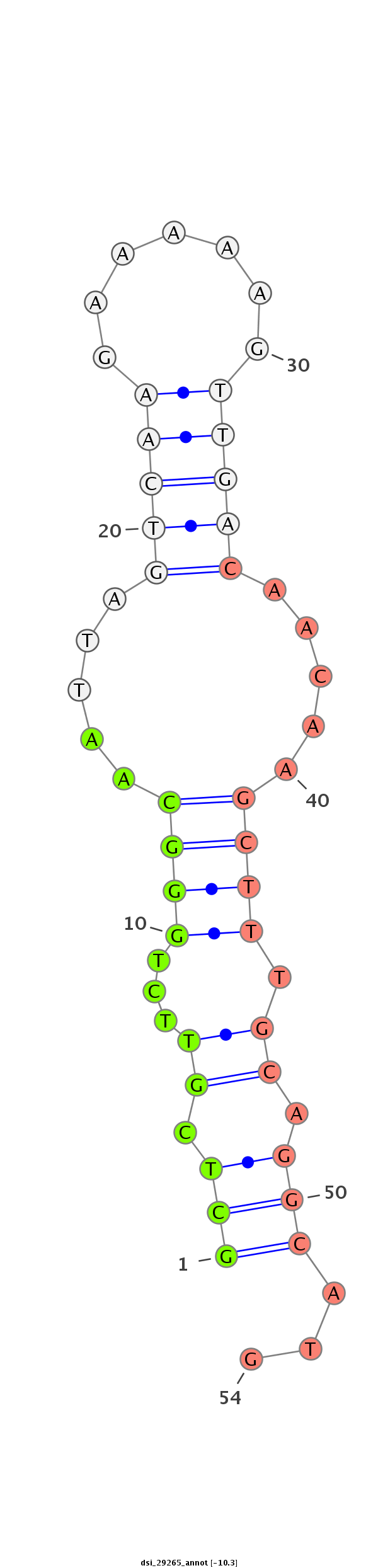


ID:dsi_29265 |
Coordinate:2r:5129090-5129239 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -9.5 |
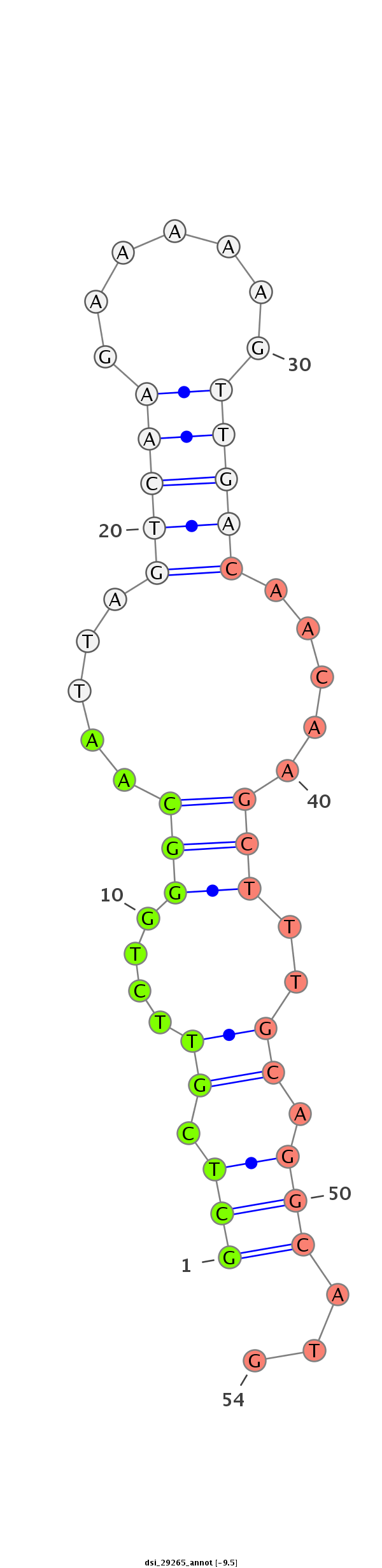 |
CDS [2r_5129240_5129460_+]; exon [2r_5129240_5129460_+]; intron [2r_5123224_5129239_+]
No Repeatable elements found
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CCATAGGCATCACAACAAACTTGTTGCAATGGCCTTCAGTGCACTCAGGAGCTGGCACTTGAGGCGACACTTTGGCCAACTTCAAAAACTGCTCGTTCTGGGCAATTAGTCAAGAAAAAGTTGACAACAAGCTTTGCAGGCATGCAGGCAACGCTAAGAGGGAGAAAACTCTCTAATTTTCCCTTTAATCCTCCACGCAGACATCATCCACGGAAAGCTGTTCCAGAACACACGCATTTTGGAGGTGGAC ******************************************************************************************(((.((...((((.....(((((.......))))).....)))).)).)))...********************************************************************************************************** |
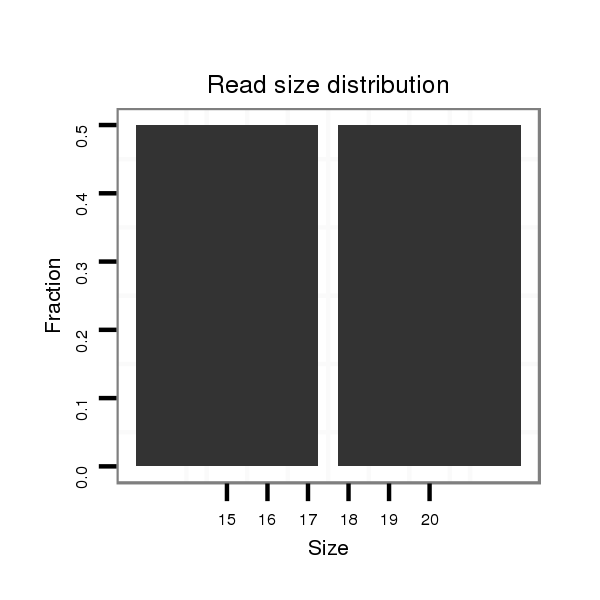
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
O001 Testis |
SRR553485 Chicharo_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
SRR902009 testis |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................GCTCGTTCTGGGCAA................................................................................................................................................. | 15 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................TCATCCACGGAAAGCTGTTCCAGAACA.................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................TCCAGAACACACGCATTTTGGAGGTGGA. | 28 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................TATGCGGACATCATCCACGGAAAGC................................ | 25 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................................CAACAAGCTTTGCAGGCATG.......................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................GCTCGTTCTGGGTAAT................................................................................................................................................ | 16 | 1 | 12 | 0.25 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................GCTCGTTCTGGGTAATTC.............................................................................................................................................. | 18 | 2 | 10 | 0.20 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................GCTCGTTCTGGGCAAATA.............................................................................................................................................. | 18 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................AAAGTGCTCGTTCTGGGTA.................................................................................................................................................. | 19 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................CTAGAAGTGCTCGTTCTGGG.................................................................................................................................................... | 20 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................CTGCTCGTTCTGGGTA.................................................................................................................................................. | 16 | 1 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................GCTCGTTCTGGGTAATTG.............................................................................................................................................. | 18 | 2 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................TAGCAACTAGTCAAGAAAAA................................................................................................................................... | 20 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................AAAATGTGCTCGTTCTGGGT................................................................................................................................................... | 20 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GGTATCCGTAGTGTTGTTTGAACAACGTTACCGGAAGTCACGTGAGTCCTCGACCGTGAACTCCGCTGTGAAACCGGTTGAAGTTTTTGACGAGCAAGACCCGTTAATCAGTTCTTTTTCAACTGTTGTTCGAAACGTCCGTACGTCCGTTGCGATTCTCCCTCTTTTGAGAGATTAAAAGGGAAATTAGGAGGTGCGTCTGTAGTAGGTGCCTTTCGACAAGGTCTTGTGTGCGTAAAACCTCCACCTG
**********************************************************************************************************(((.((...((((.....(((((.......))))).....)))).)).)))...****************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
GSM343915 embryo |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|
| ............................................................................GTTGAAGTTTCTGATGAGCAC......................................................................................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ...............................................................................................................................................................................................AGGAGCGTATGTAGTAGGCGC...................................... | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ....................................................................................................................................................................................GGGCAATTAGAAGGCGCGT................................................... | 19 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 |
| ..................................................................................TTTTTTGACGAGAAAGAC...................................................................................................................................................... | 18 | 2 | 15 | 0.07 | 1 | 0 | 1 | 0 |
| ......................................................................................................................................................................TTGTGAGATTAAAAGAG................................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ......................................................................................................................................................................................GCAATTAGAAGGCGCGTC.................................................. | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
Generated: 04/23/2015 at 09:53 AM