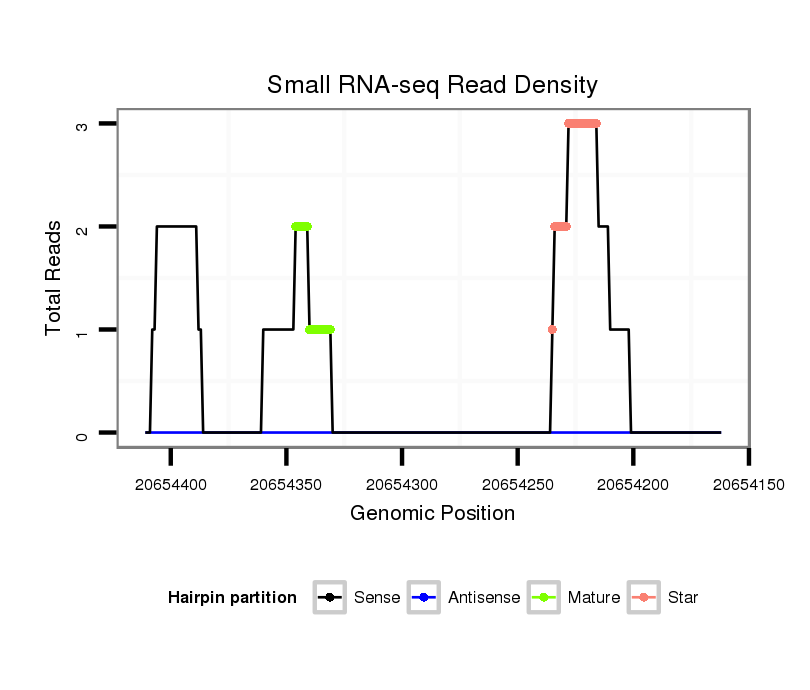
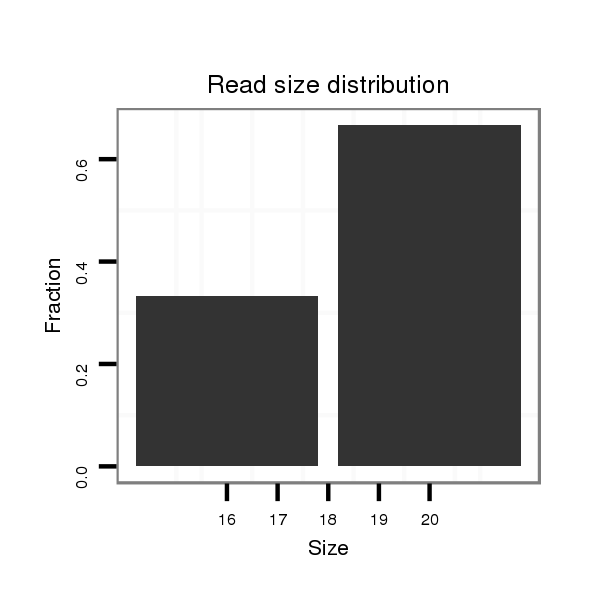
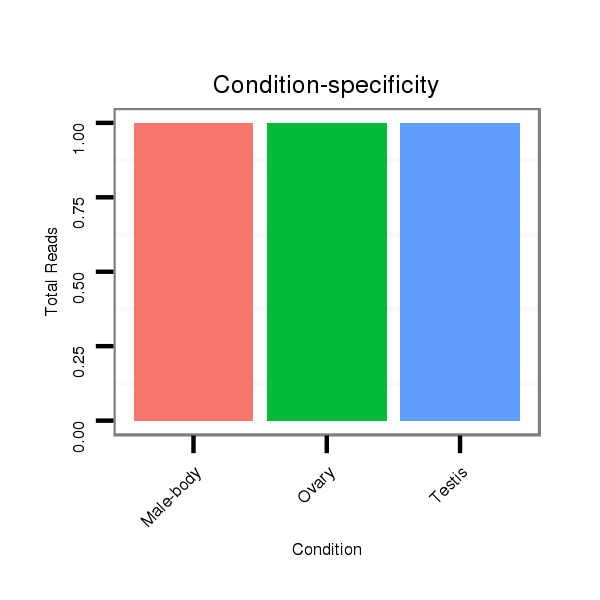
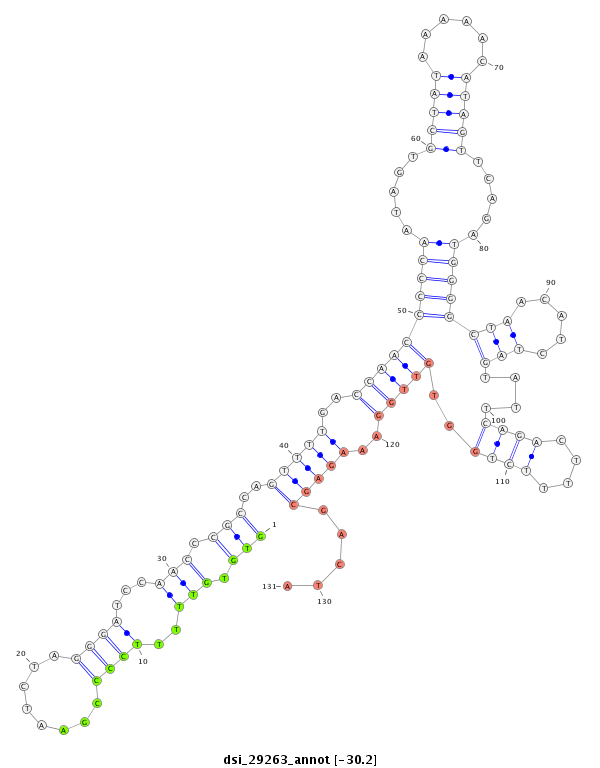
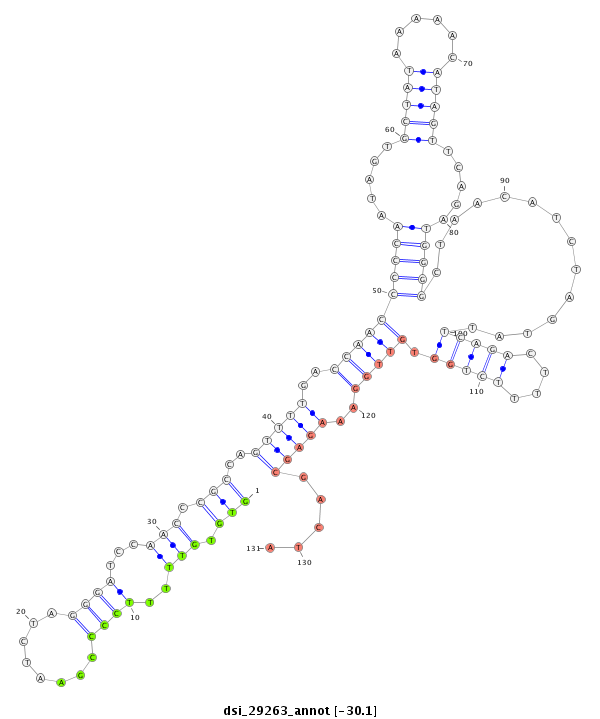
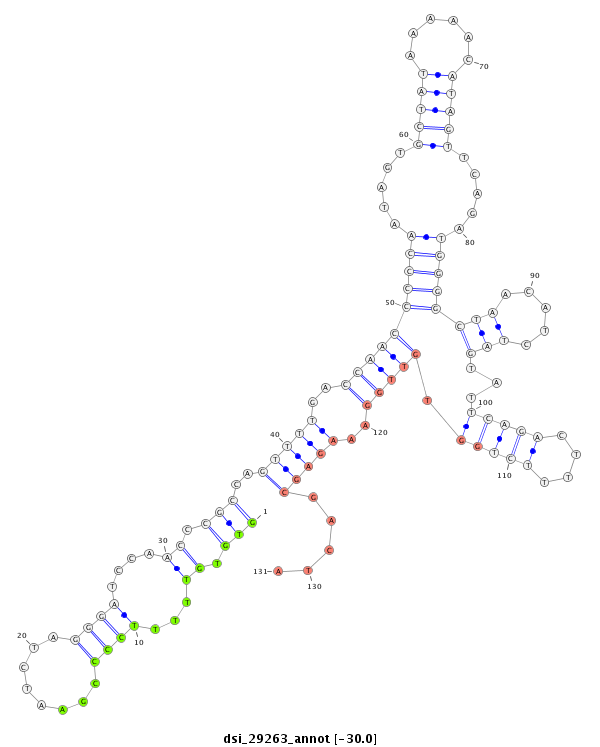
| droSim2 |
3r:20654162-20654411 - |
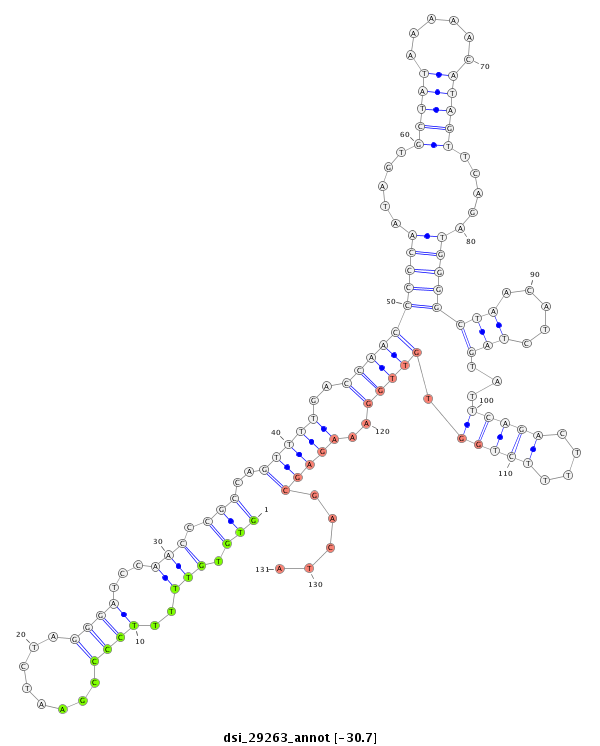
dsi_29263 |
GGA---TTGCAGTCGA-----------------TCTATCAGT-----------ACGCTCCGT----TTACCAATTAAGGAAAAAGGTTTGCTACCGCTGTG-TGTGTTT--------------------------------------TTCCC-----CGAATCTAGGGATCCAACCCGCCAGTTTTGACCAACCC---CCA--------------------------------------------------------------------ATAGTGCTATAAAAACATAGTT--CAGATGGG-GCTAACATCTAGTATTCAGACTTTTCTGGTGTTG---GAAAGAGCGACT-------------------------------AAGAG-TGAAATTGGCTTATAA-----G----------------------AAAG-----TGCATTGAAAATTG----------------------TT--CAAA------AATCAATAC |
| droSec2 |
scaffold_33:398173-398440 - |
|
GGA---TTGCAGTCGA-----------------TCTATCAGT-----------ACGCTCCGT----TTACAAATTAAGGAAAAAGGTTTGCTACCGCTGTG-TGTGTTTTCGCA-------------------------GTGGGCCATTCCC-----CGAATCTAGGGATCCAACCCGCCAGTTTTGACCAACCC---CCA--------------------------------------------------------------------ATAGTGCTATAAAAACATGGTT--CAGATGGG-ACTGACATCTAGTATTCAGACTTTTCTGGTGTTG---CAAAGAGCGACT-------------------------------AAGAG-TGAAATTAGCTTATAAGCAAAG----------------------AAAG-----TGCATTGAAAATTG----------------------TT--CAAA------AATCAATAC |
| dm3 |
chr3R:21139265-21139532 - |
|
GGA---TTGCAGTCGA-----------------TCTATCAGT-----------GCGCTCCGT----TTACCAATTAAGGAAAAAGGTTTGCCACCGCTGTA-TGTGTTTTCGCA-------------------------TTGGGCCATTCAC-----CGAATCTAGGGATCCGACCCGCCAGTTTTGACCAACCC---CCA--------------------------------------------------------------------ATAGTGCTATAAAAACATAGTT--CAGATGGG-ACTAACATCTAGTATTCAAACTTTTCTTGTGGTG---GAAAGAGCGACT-------------------------------AAGAG-TGAAATTAGCTTATAAGCTAAG----------------------AAAG-----TGCATTGAAAATTG----------------------TT--CAAA------AATCAAAAC |
| droEre2 |
scaffold_4820:6904460-6904690 + |
|
GGA---TTGCAGTCGA-----------------TCTATCAGC-----------CCGCTCCGT----TTACCAATTAAGGAAAAAGGTTTGCTACCAATGTG-TGTGTTTTCGCA--------------------------------------------GAATTGGGGGTTCCAAA----------------------G--------------------------------------------------------------------GACAAAGTGCTATAAAAACATGGCT--CGGATGGG-ACTGACATCTAGTATTCAGACTGTTATTGTGGTG---GACTGAGCGACT-------------------------------AAGAG-TGAAATTGA----AAAACAAAG----------------------AAAG-----TGCATGGAAAATTC----------------------TT--AAAA------AAACAAAAC |
| droYak3 |
3R:25349206-25349456 + |
|
GGA---TTGCAGTCGA-----------------TCTATTAGC-----------CCGCTCCGT----TTACCAATTAAGGAAAAAGGTTTGCTGCCA---------------------------------------------------TTCCC-----AGAACGGGGGTATCCAATCTGCCAGTTTTGACCAACAA---CTA----------------------------------------------------------TCGG--GAACAAAGTGCTATAAAAACATGGAT--GCGATGGG-GTTAACTTCTAGTATTCAAACTGTTATTGTGGTG---GATTGAGCGATT-------------------------------GGAAG-TGAAATTAGTTTATAAGCAAAG----------------------AAAG-----TGCATGGAAAATTG----------------------TT--GAAA------ATCCAAAAC |
| droEug1 |
scf7180000409692:237903-238205 + |
|
GGA---CTGCAGTCGACCGA-------------TCCATCAGC-ACG-ATTC--CCGCTCCGT----TTACCTATTAAGGAA-AAGGTTTGCTACCGTTGAT-TGTGTTTTCCCA-------------------------GTGACACAT----ATTCCC----CGAGGGATCCAATCTCTCAGTTTAGACCAACAG---CTA----------------------------------------------------------TCGG---CTCATATTGCTATAAATATGTAGCT--CAGATTGG-TCTAAAATCTAGTATACCAACTGTTATTATAGTG---GAAAGGGTGAAT-------------------------------CTAAG-TGGAATTAGTTAATGAAACA--------------------------AGTTTGGTGAATAGAA-GTTATTGAAAGTGAAGTTATTTGTTCCT--TAAA------AACCAATA- |
| droBia1 |
scf7180000302136:1959077-1959340 - |
|
GGG---CTGCAGTCGA-----------------TCTATCAGA-----------ACGCTC-GT----TTACCTATTAAGGAAGAAGGTTGGCTACCGATCTG-TT--CTT----------------------------------------TCC-----CGATTCCACGAATCCAATCTCCCAGTTTTGACCATCAAAAG-------------------------------------------------------------T-------GAAGAGCGCTATAAAACCATAGCT--CAGATTGG-GCCAACCTCTAGTTTACCAACTAGTATCATAGTG---AAAAGGTCGTTT-------------------------------TATAG-TGAAATTCGTC---GCACCT--------------------------ATATTAGTGAATAAAT-CATATTTTAAGTGACGTT----------TTCGTA------TCCTAAAGC |
| droTak1 |
scf7180000413457:74203-74491 + |
|
GGA---CTGCAGTCGA-----------------TCTATCAGA-----------ACGCTCCGT----TTACCTATTAAGGAAAAAGGTTTGCTACCGCTGTG-TA--CTTTCCCA-------------------------GTGACACATTCCC-----TGGTTCGAGGGATTCAATCTCTTAGATTTGATTATCAGAAA-------------------------------------------------------------TG-------AAAATTTCTATAAAAACGTAGCT--CAAATTGG-CCTAAAATCTAGTATAACAACATATATTGTGGTG---GAAAGAACGCAT-------------------------------TATAT-TTGAAATAGTTTAAAAACCA--------------------------AGTATAGTGAATAGAA-ATTATTAAAAGTGAAGTT----------TTCGCAGCCCTCAGTCAATAT |
| droEle1 |
scf7180000491212:505779-506054 - |
|
GGA---CTGCAGCCGATCTA-------------TCTATCAGC-ACG-AATCCAGCGCTTCGA----TTACCAATTTAGGAAAAAGGTTTGCTACCGATGTG-TA--TTTTCCCAA----GATTGGTGACACATACACAT-------------AGTCCCGATTCGAGGCAACCAATCTCCCAGTT-CGACCCAC----------------------------------------------------------------------------AGAGTGCTATAAAAATGTATCT--GAGATTGGAAGTAACAGCTAGTATAACAAATAACATTAAGGTGT--TAAGGGGCAGTT-------------------------------CAAAA-TAAAA------------CTT--------------------------AGTTTAGTGAAAAACA-GATACCCAAAGAGAAGTT------------------------------T |
| droRho1 |
scf7180000779365:104476-104731 + |
|
GGC---CTGCAGTCCA-----------------TCTATCAGC-ACG-AATC--CCGCTCCGA----TTACCAATTAAGGAAAAAGGTTTGCTACCGATGTG-TA--TTTTCCCAA----GATTGGCAACAC---------------------AGTCCCGATTCGAGGGAACCAATCTC-------TGACCAAC----------------------------------------------------------------------------AGAGTGCTATAAAAACGTAGCT--CGGATTGG-AGAAAC-TCTAGTATACAAAATAAAATTGTGGTG---AAAAGGGCAAAT-------------------------------TAAAG-TGAAATTCATTGACAAACTG--------------------------AGTTCAATGAGAAAGA-GTTC----------------------T---C-----------T---CAC |
| droFic1 |
scf7180000454055:1056116-1056365 - |
|
GTG---TTGCAGTCGA-----------------TCTATCAGC-GCG-ATTC--CCGCTCCGT----TTGCCAATTAAGGTAAAAGGTTTGCTGG----------------------------------------------TGACACATTTCC-----CGATTCTTCGGAACCAATCTCATGG-TTTGACCATCAG---CTA----------------------------------------------------------TCAGATGGACTGAGTGCTATAAAAACGGTGTT--AGAATCGG-AGCAAGATCTAGTATATCAACTAGTATTGTGGTATTGCAAACGTGAAAT-------------------------------TAAAG-TGAAAACAT-TTAGTGGCTA---------------------------------------------------------AGTT----------CGCGAG------TCCCAATAA |
| droKik1 |
scf7180000302355:1378071-1378290 + |
|
GAA---CTGCAGTCAC-----------------CC--ACGGA-----------GCGCTCCACTAGCTTACCAATTAAGGAGAAAGGTTTGTCACCATT----------------------------------------------------------------TCGGGGAACCAATCTCTTCGTTTTGACCAACCAAGT-------------------------------------------------------------TC-------GGAACTGATATAAAAACCCATTT--TCCATCAT-CGGAACTTTTAGTAATTAAATAATTGTGGCGGTG---AA-GGAGCT----------------------------------GTGAA-TTAAATTAT----CA---------------------------------------------AA-ATTATT-AGTGTGAAGTT----------CTTGTA------------TCC |
| droAna3 |
scaffold_13340:1339061-1339311 - |
|
GGACCACTGCAGTCGA-----------------TCCATCAGCC-GGATA----CCGCTCCAC-----TTCCAATTAAGGAAAAAGGTTCGACAAGGGTTCGGTGAATCACC-----------------------------------------------ACATCGGGATATCTT-GGTAGC--------------G---CCA---------T----------------------------------------------------------GGATCACTATAAAAGG-CGGTA-GCAAATGGC-AGTTAACTTTATTATTGACTCTAAT-TTTTGGTA---GGA------A--TCATTCGGGTCCTTA-----CTCACAATTTA-----------------------------------TAGGAAAACAAGGAAAAAG-----TGTTTTGATTATTA-----------------GGT--TT--CTTA------AATCAA--- |
| droBip1 |
scf7180000396712:486038-486291 + |
|
GGACCACTGCAGTCGA-----------------TCCATCAGCC-GGATA----CCGCTCCAC-----TTCCAATTAAGGAAAAAGGTTGGCCAAGGGTTCGGTG------------------------------------------AATCAC-----CAAATCGGGGTATCTTAGGTAGC--------------G---CCA---------T----------------------------------------------------------GGGTCATTATAAAAG--GCGCTAACAAATGAC-AGTTAATTTTAGTAATGACACTACT-TTGTGATG---TACATATAAA--TCGTTCGAGTCCTTAACGTCCTTGC------------------------------------------------ACAAATAAAAGG-----TGTGGTGAAATTTT----------------ATAT--TT--TAAA------AATGAAAGA |
| dp5 |
2:20529711-20529789 + |
|
GGA---CTGCAGTCAATCGAGAGCTGCT--CGAATTATCAGGCACG-TAGC--TCCCTCCAT----TCACCAATAAAGGAAAAAGGTTTGC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droPer2 |
scaffold_3:3283826-3284142 + |
|
GGA---CTGCAGTCAATCGAGAGCTGCT--CGAATTATCAGGCACG-TAGC--TCCCTCCAT----TCACCAATTAAGGAAAAAGGTTTGCA-CGTCCAAA-AATCTCTTCGTAGCTGG--------------------GTGACACATTCGC-----GATTTCCACGGAA----------------------AGT---TCTTTAGTGCCTTATACGGTCAATACTGTGTGGAGTGTGCTGTTTGCTGGCAGTCCAGGTGTG-------AAAA----TATATAAATAAGGCT------------------------------------------------------ACGAAT-------------------------------AAGACCCAAAATTTAGTCATAGGCCAAGGTTTTGGAGTGAAGAT-AATACATAGTTCTGTGCGTTA-----TT--------ATATTTAATTATCCTT---ATA------AAAGGATAA |
| droWil2 |
scf2_1100000004943:13481414-13481442 + |
|
-------------------------------------------------------------T----TGGCCAATTAAGGAAAAAGGTTTGCCAC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droVir3 |
scaffold_13047:2260498-2260568 + |
|
GAA---CTGCACTCGATTGTG-----TTAGCAATTTATCACTC--G-AG----TCTCTCATT----TTGTCAATTAAGGAAAGAGGTTTG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droMoj3 |
scaffold_6540:22647883-22647958 - |
|
GAA---CTGCATTCGATTGTG-----TGAACAATTCATCAT-----------------TCG-----CCGTTAATTAAGGAAAAAGGTTTGTTGTG-------CCTATCTCTCC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droGri2 |
scaffold_14906:9111642-9111712 + |
|
GAA---CTGCATTCGATCGTG-----CTAACAATTTATCCATCGA---G----CCTCTCATT----TTGTCAATTAAGGAAAAAGGTTTG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |