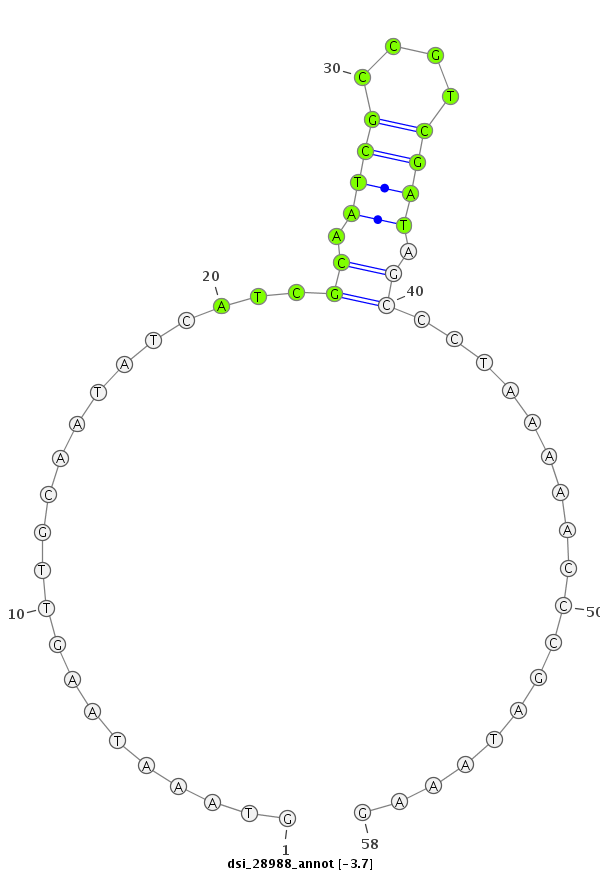
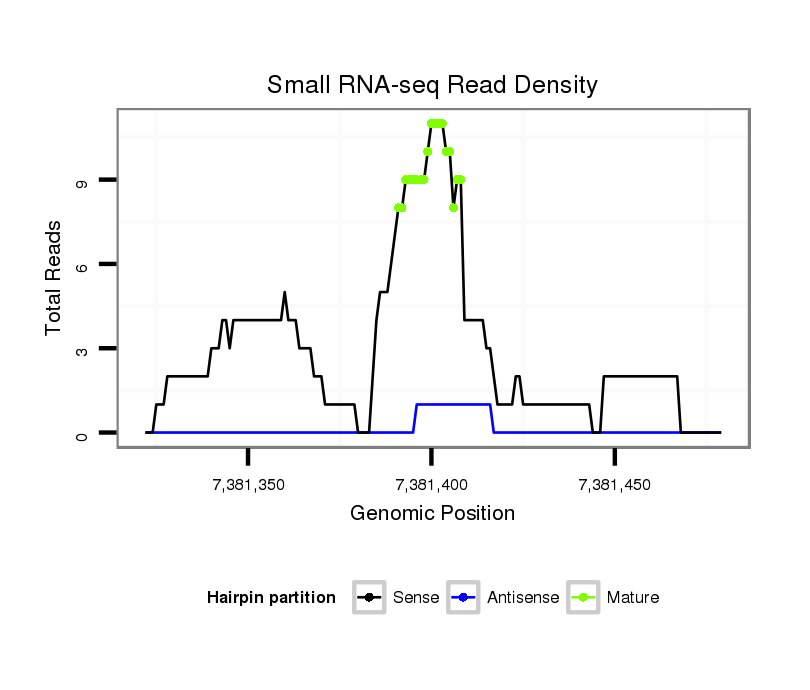
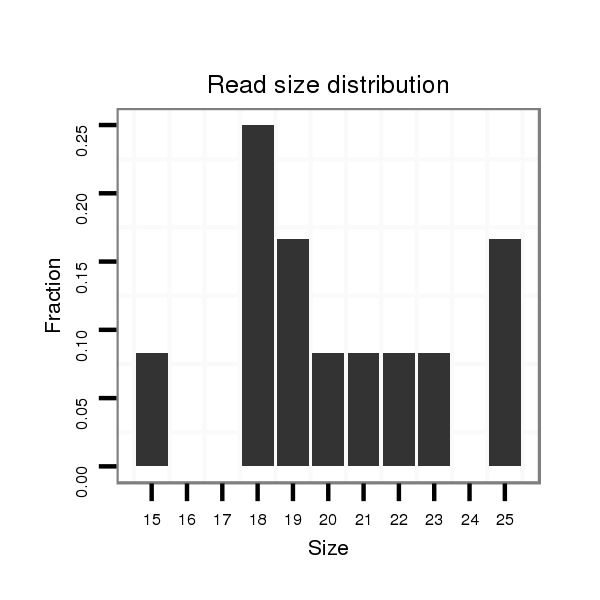
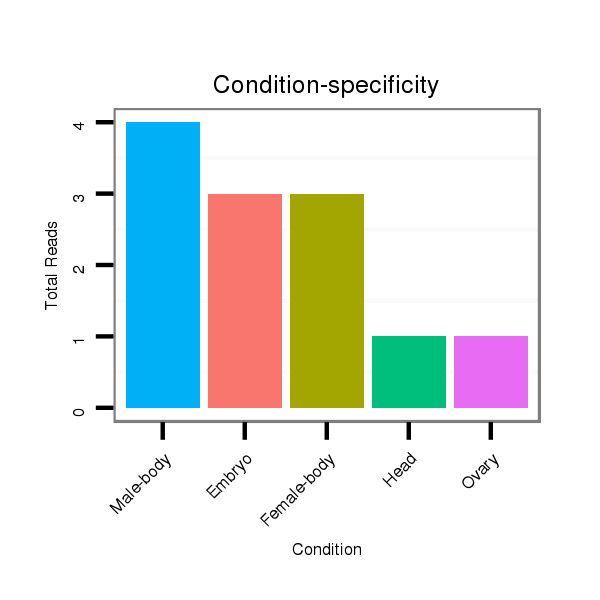
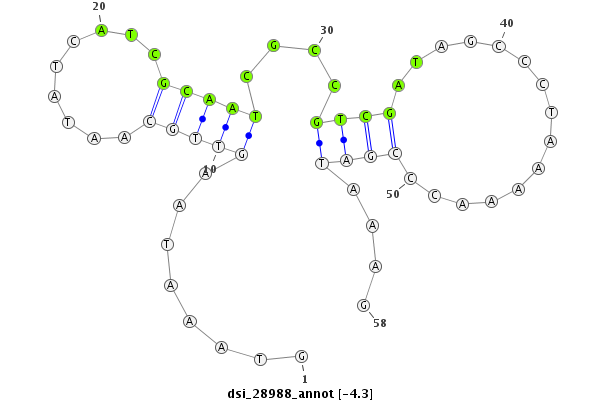
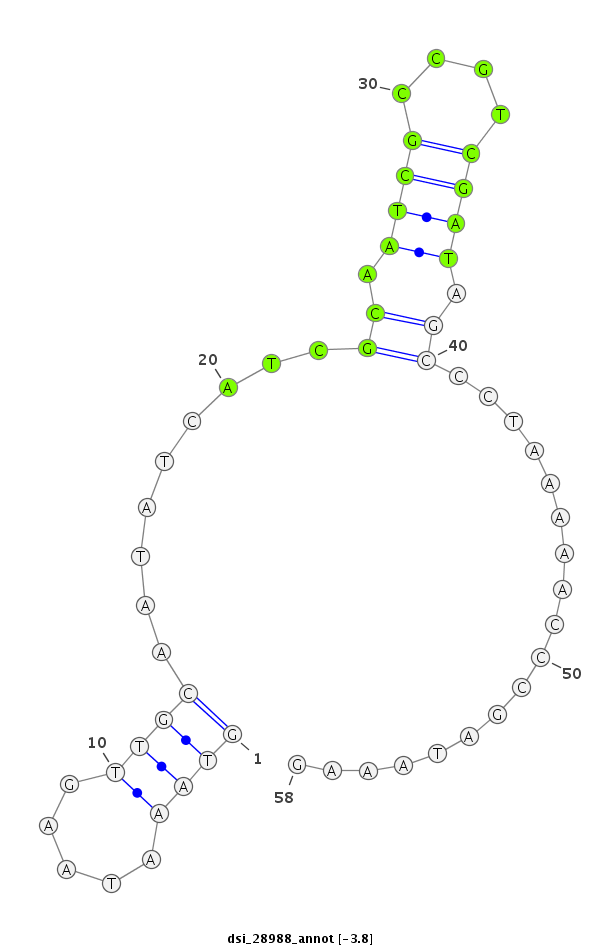
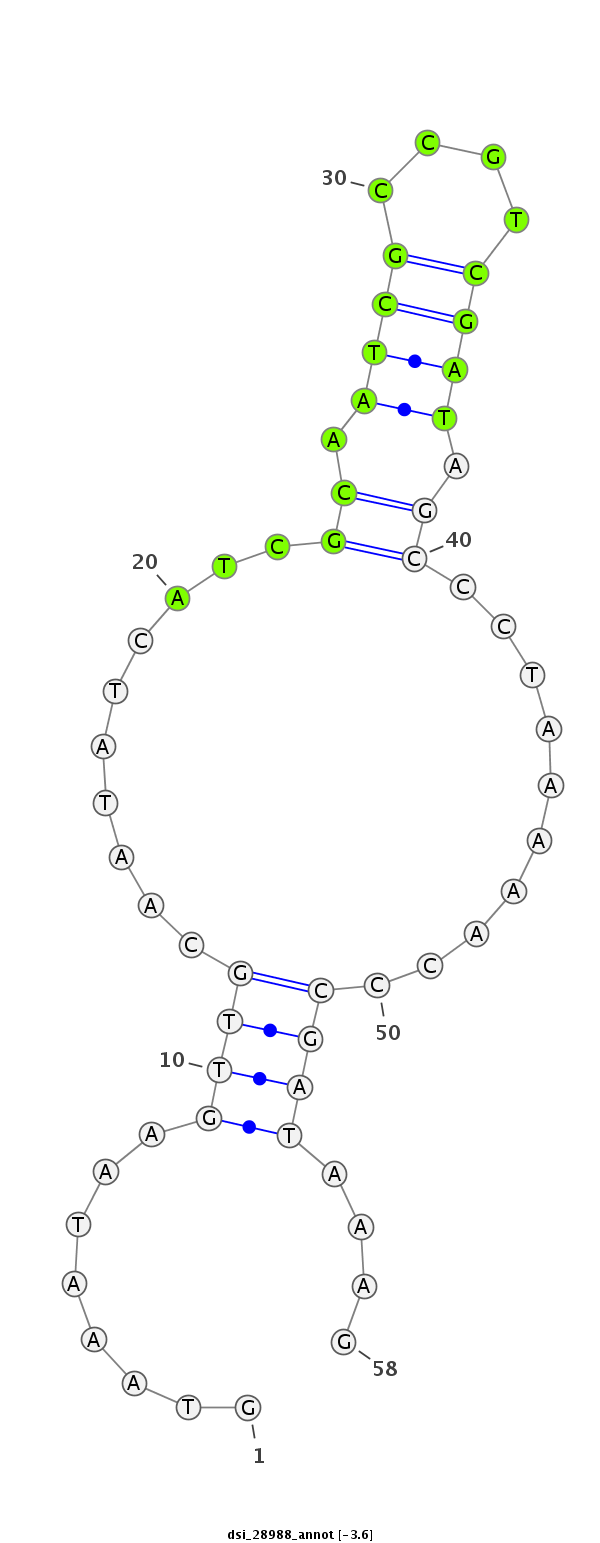
ID:dsi_28988 |
Coordinate:3l:7381372-7381429 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -4.3 | -3.8 | -3.6 |
 |
 |
 |
five_prime_UTR [3l_7381332_7381718_+]; exon [3l_7381332_7381718_+]
No Repeatable elements found
| ----------#################################################################################################################################################### AAAGCAGAGAAAGAAATAATAATTCGTGTTTTGTGTGCAGAGTGCAAAGGGTAAATAAGTTGCAATATCATCGCAATCGCCGTCGATAGCCCTAAAAACCCGATAAAGTGCCAGGATTTTTGGATATCCATATCGTGCGCGCAGCTCTACTATTTTCC **************************************************......................((.((((....)))).))..................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
M025 embryo |
M053 female body |
O002 Head |
SRR618934 dsim w501 ovaries |
GSM343915 embryo |
M023 head |
O001 Testis |
SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................ATCCATATCGTGCGCGCAGCT............ | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................AGAGTGCAAAGGGTAAATAA.................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................ATCGCAATCGCCGTCGAT....................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......GAGAAAGAAATAATAATTC..................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................CGCCGTCGATAGCCCTAA............................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................AATATCATCGCAATCGCCGTCGAC....................................................................... | 24 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................AATATCATCGCAATCGCCGTC.......................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................CGCAATCGCCGTCGATAGCCCTAAA.............................................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................ATAATTCGTGTTTTGTGTGCA....................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................GTGTTTTGTGTGCAGAGTGCAAAG............................................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................GATAAAGTGCCAGGATTTTTG.................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................GCCGTCGATAGCCCT................................................................. | 15 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................CGTGTTTTGTGTGCAGAG.................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................ATATCATCGCAATCGCCGTCGAT....................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................CAATATCATCGCAATCGCCGTC.......................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................TTGTGTGCAGAGTGCAAAGGGTAAAC...................................................................................................... | 26 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................ATCCATATCGTGCGCGCAGCTCTACG....... | 26 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................CAATATCATCGCAATCGCCGTCGAT....................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................TCATCGCAATCGCCGTCGAT....................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................ATAGCCCTAAAAACCCGA....................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...GCAGAGAAAGAAATAATAAT....................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................CATCGCAATCGCCGTCGAT....................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................AATATCATCGCAATCGCCG............................................................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................ATTCGTGTTTTGTGTGCAGAGTGCA................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................CCGTCGATCGCCCTA................................................................ | 15 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TTTCGTCTCTTTCTTTATTATTAAGCACAAAACACACGTCTCACGTTTCCCATTTATTCAACGTTATAGTAGCGTTAGCGGCAGCTATCGGGATTTTTGGGCTATTTCACGGTCCTAAAAACCTATAGGTATAGCACGCGCGTCGAGATGATAAAAGG
**************************************************......................((.((((....)))).))..................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
SRR902009 testis |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|
| ..................AAATAAGCACAAAACACACGTCT..................................................................................................................... | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................ACGCGCGTCGATATGATAAAA.. | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ..........................................................................TTAGCGGCAGCTATCGGGATT............................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ..................................................................................ATCTCTCGGGATTTTTGGACT....................................................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................TCGCGCGCACGTCGAGATG........ | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................TCGCGCGCACGTCGAGAT......... | 18 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 |
Generated: 04/24/2015 at 06:50 AM