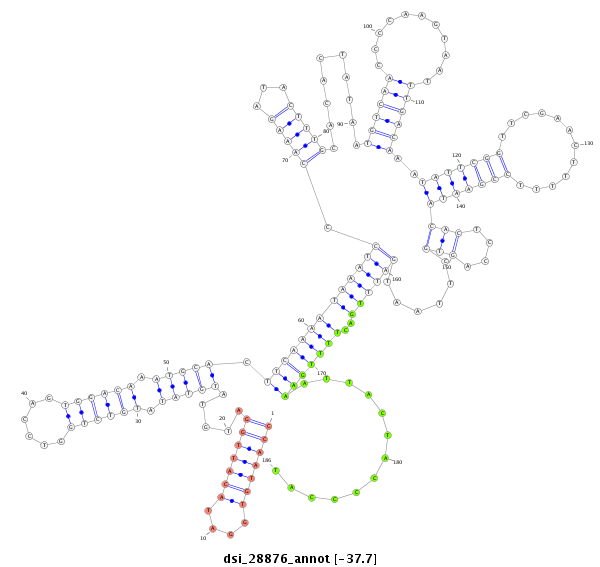
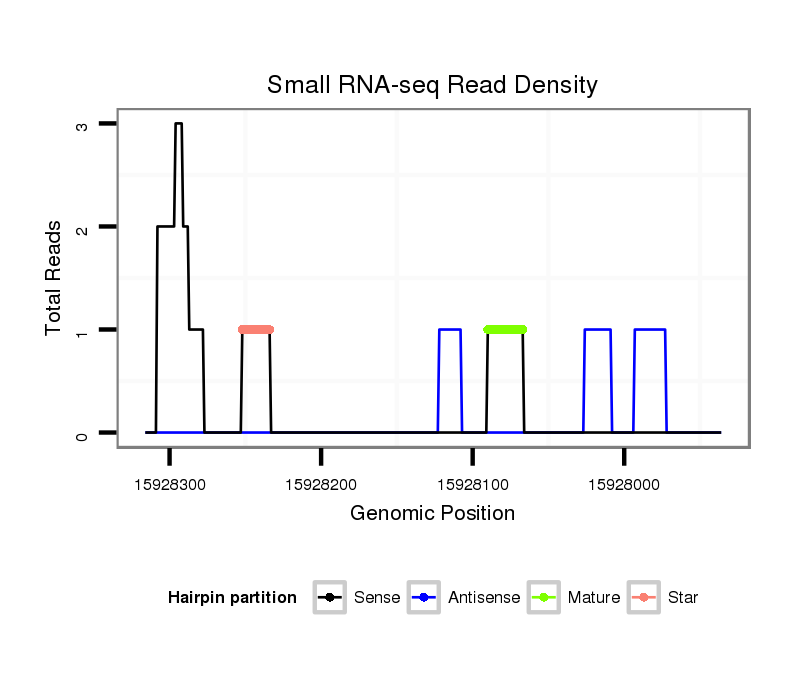

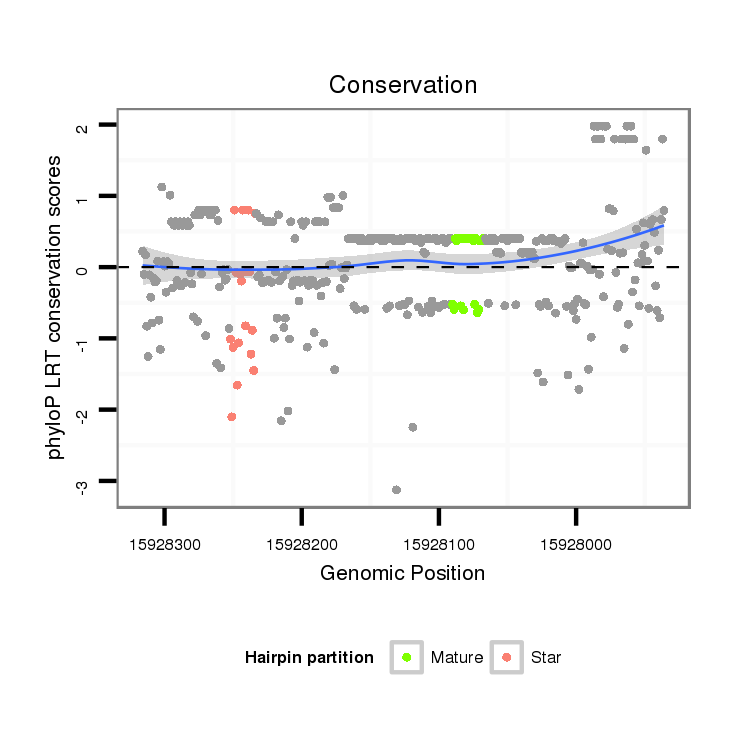
ID:dsi_28876 |
Coordinate:3r:15927986-15928266 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -37.1 | -36.9 | -36.9 |
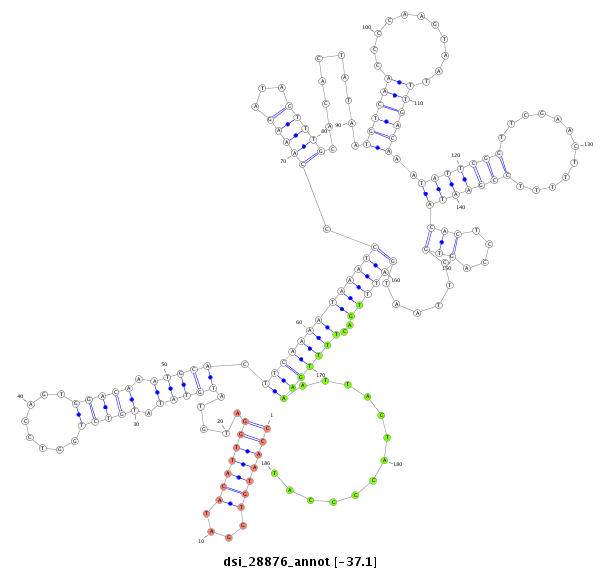 |
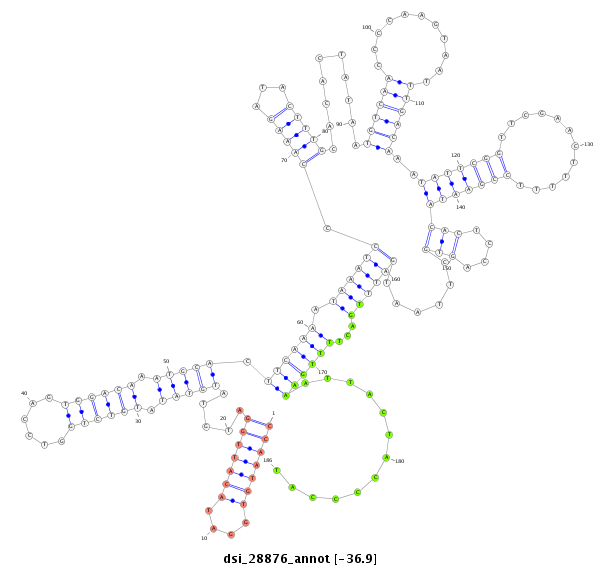 |
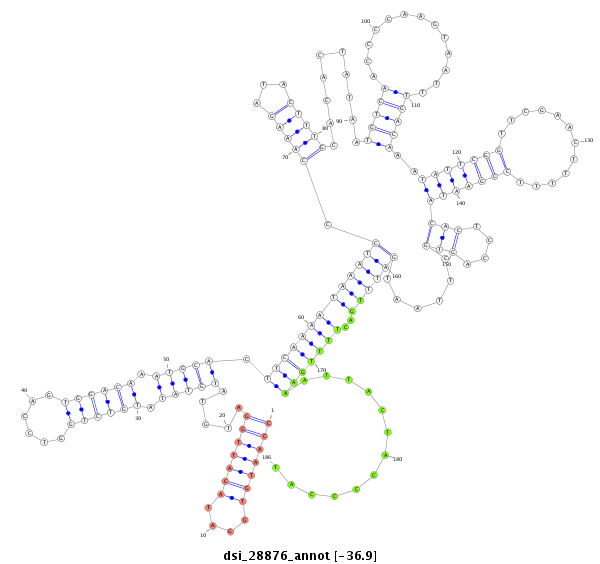 |
exon [3r_15928267_15928354_-]; CDS [3r_15928267_15928354_-]; exon [3r_15927914_15927985_-]; CDS [3r_15927914_15927985_-]; intron [3r_15927986_15928266_-]
No Repeatable elements found
| ##################################################-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TGTACACTTCTGCCGACGAGAAGGATCATGACGAGGCTCCCAGGATTAAGGTAACTCAATGCTTCCAATGTGGATACATTGGATGTATGTATATGTCTGGTCCAGTGGACAAATGCACTTCAAAATAAATCCCAAAGATACTTTGCACACTATAATGTCAACCCCAAGTAATTTGACAAATATTCGGTTCGAACTTTTTCCGAATACACTCCAGTGCTTAATGATTTGACTTTTGAAATTACTACCCCATTCTTCTACATTCATATGTAAATCAAATGTTACTTATTATAGAACACTAATGCTTACACTCAAAAATATATTTTTTATGCAGATCCAGAAGATTTCTGGCGATGATGATGGCGTCTTGGATAATGCCAACGA ****************************************************************(((((((....))))))).....(((((.((((((......)))))).))))).(((((((((((((.(((((...)))))..........((((((...........))))))..(((((((............)))))))(((....)))......))))))..))))))).............*********************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
SRR902009 testis |
SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
SRR553486 Makindu_3 day-old ovaries |
M024 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............ACGAGACGGATCATG............................................................................................................................................................................................................................................................................................................................................................... | 15 | 1 | 4 | 1.25 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................AGAAGGATCATGACGAGGCTTT..................................................................................................................................................................................................................................................................................................................................................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........TCTGCCGACGAGAAGGATCAT................................................................................................................................................................................................................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........TCTGCCGACGAGAAGGA.................................................................................................................................................................................................................................................................................................................................................................... | 17 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................................................................TGACTTTTGAAATTACTACCCCAT................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................AAGGATCATGACGAGGCTC...................................................................................................................................................................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................CCAATGTGGATACATTGGA.......................................................................................................................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................GATGCAGATCCGGAAGATTAC.................................... | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................TGCTTGATGATTTGA........................................................................................................................................................ | 15 | 1 | 18 | 0.17 | 3 | 0 | 0 | 2 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................CTGATGGCGTGTGGGATAA......... | 19 | 3 | 18 | 0.11 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| .......................................CCAGGATGAAGGTAA....................................................................................................................................................................................................................................................................................................................................... | 15 | 1 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................TATGCAGATCCGGAACA........................................ | 17 | 2 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................TGCTTAATGATTCGA........................................................................................................................................................ | 15 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................GAAGATGATGGCGTC................. | 15 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................TTCCAACGTGGATACCCTG............................................................................................................................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
ACATGTGAAGACGGCTGCTCTTCCTAGTACTGCTCCGAGGGTCCTAATTCCATTGAGTTACGAAGGTTACACCTATGTAACCTACATACATATACAGACCAGGTCACCTGTTTACGTGAAGTTTTATTTAGGGTTTCTATGAAACGTGTGATATTACAGTTGGGGTTCATTAAACTGTTTATAAGCCAAGCTTGAAAAAGGCTTATGTGAGGTCACGAATTACTAAACTGAAAACTTTAATGATGGGGTAAGAAGATGTAAGTATACATTTAGTTTACAATGAATAATATCTTGTGATTACGAATGTGAGTTTTTATATAAAAAATACGTCTAGGTCTTCTAAAGACCGCTACTACTACCGCAGAACCTATTACGGTTGCT
***********************************************************************************************************************************(((((((....))))))).....(((((.((((((......)))))).))))).(((((((((((((.(((((...)))))..........((((((...........))))))..(((((((............)))))))(((....)))......))))))..))))))).............**************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR902009 testis |
SRR618934 dsim w501 ovaries |
O002 Head |
SRR553488 RT_0-2 hours eggs |
SRR902008 ovaries |
SRR553485 Chicharo_3 day-old ovaries |
GSM343915 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................AGTGATGAAGGTTACACCCAT................................................................................................................................................................................................................................................................................................................. | 21 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................AAAAAGGCTTATGTG............................................................................................................................................................................ | 15 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............GCTGCTGTTCCTAGTACTGGT........................................................................................................................................................................................................................................................................................................................................................... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................AATACGTCTAGGTCTTCTAAA..................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................CTTGTGATTACGAATGTG......................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................ACGTTAAGTTTTATTTAGAGTAT..................................................................................................................................................................................................................................................... | 23 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................TACTGCTCCGAGGTTCGT................................................................................................................................................................................................................................................................................................................................................ | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................AAGGCTTAAGTGGGGTCA...................................................................................................................................................................... | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................GGCTAATAAGATGTAAGTAGA................................................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................ACATTTAGTTTACAA..................................................................................................... | 15 | 0 | 6 | 0.33 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................TGCCGCTACTAGTACCGCAGA................ | 21 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................ATTTATTCACGAAGGTTACACCT................................................................................................................................................................................................................................................................................................................... | 23 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................TACGTCTAGGCCTTGTA....................................... | 17 | 2 | 20 | 0.25 | 5 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................AGGCTTATGTGTGGTC....................................................................................................................................................................... | 16 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................AAAACGTGCATGATGGGGTA................................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................AAAAGGCTTATGTGAT.......................................................................................................................................................................... | 16 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................ACAAGGCTTAAGTGGGGTCA...................................................................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................ACATTTAGTTTACAAT.................................................................................................... | 16 | 0 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................AAAAAAAACATCTTGGTCTTCT........................................ | 22 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................TTAGTTTACAATGAAAAA.............................................................................................. | 18 | 1 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................................................................................................................................................................................................................................................TATCTTGTGATTAGGAATT........................................................................... | 19 | 2 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................ATACGTTTAGTTTACAAT.................................................................................................... | 18 | 1 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............CTGCTCTGCCTAGTACC.............................................................................................................................................................................................................................................................................................................................................................. | 17 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................AGTTAGGAAGGATACA...................................................................................................................................................................................................................................................................................................................... | 16 | 2 | 19 | 0.11 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................CAAGGCTTAAGTGGGGTCA...................................................................................................................................................................... | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................ACTGCTCCGAGGTTCGT................................................................................................................................................................................................................................................................................................................................................ | 17 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................AGGGTCCTACTTCCCGTGAG.................................................................................................................................................................................................................................................................................................................................... | 20 | 3 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................AGAGTGAACAAGGCTTATGT............................................................................................................................................................................. | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................AGGCTTAAGTGGGGTCA...................................................................................................................................................................... | 17 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................AAAATACTTCTAGGTCGT.......................................... | 18 | 2 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................AACTACGTCTAGGCCTTGTA....................................... | 20 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................TGAGTTTCGAAGGTTAT....................................................................................................................................................................................................................................................................................................................... | 17 | 2 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................ACATTTAGTTGACAA..................................................................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................TACTATCTCGTGATCACG............................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................AAGAAGAAGTAAGTA..................................................................................................................... | 15 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................AGGTCACGTATTACTAG........................................................................................................................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 3r:15927936-15928316 - | dsi_28876 | TGTACACTTCTGCCGACGAGA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGGATCATGACGAGGCTCCCAGGATTAAGGTAACTCAATGCTTCCAATGTGGATACATTGGATGTATGTATATGTCTG------------------------------------------------GT--------CCAGTGGACAAATGCACTTCAAAATAAATCCCAAAGAT-----------------------------------------------------------------------------------------------ACTTTG-------------------------------------CACACTATAATGTCAACCCCAAGTAATTTGACAAATATTCGGTTCGAACTTTTTCCGAATACAC--TCCAGTGCTTAATGATTTGACTTTTGAAATTACTACCCCAT-TCTTCT-------ACATTCATATGTAAATCAAATGTTACTTATTA--TAGAACACTAATGCTTACACTCAA----AAATATATTTTTTATGCAGATCCAGAAGATTTCTGGCGATGA---TGATGGCGTCTTGGATAATGCCAACGA |
| droSec2 | scaffold_0:16796459-16796852 - | TGTACACTTCTGCCGACGAGA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGGATCATGACGAGGCTCCCAGGATTAAGGTAATTCGATGCTTCCAATGTGGATACATTGGATGTATGTATATGTCTG------------------------------------------------GTCTAGGACTCCAGTGGACAAATGCACTTCAAAATAAATCCCAAAGAT-----------------------------------------------------------------------------------------------ATTTTG-------------------------------------CACACTATAATGTCAACCCCAAGTAATTTGACAAATATTCAGTTCAAACCTTCTCCGAATACAC--TCTAGTGCTTAATGATTTGACTTTTGAAATTACTACCCCATATCTTCT-------ACATTCATATGTAAATCAAATGTTACTTATTA--TAGAACACTAATGCTTACACTCAAAAAAAAATATATTTTTTATGTAGATCCAGAAGATTTCTGGCGATGA---TGATGGCGGCTTGGATAATGCCAACGA | |
| dm3 | chr3R:5085014-5085403 + | TGTACACTTCTGCCGACGAGA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGGATCATGACGAGGCTCCTAGGATAAAGGTAATTCGATGCTTCCAATTTGGATACATTGGATGTATGTATATGTCTG------------------------------------------------GTGTAGGACGTCAGTGGACAAATGCACTTCAAAATAAATCCCAAAGAT-----------------------------------------------------------------------------------------------ACTTTG-------------------------------------CACACTATAATGTCAACCCCAAGTAATTTGACAAATATTCGGTTCGAACTTTGTCCGAATACAATATATAGTGCTTAATGATTTGACTTTTGAAATTACTAACCCATATCGTCTCATATGAACATTCATATGTAAATTAAATGTTACTTA-----TATATCGCTAATG------CTCTA----AAATATATTTTTTATGCAGATCCAGAAGATTTCGGGCGACGA---TGATGGCGGCCTGGATAATGCCAACGA | |
| droEre2 | scaffold_4770:16511047-16511412 - | TGTACACCTCTGCTGACGAGA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGATCAGGACGAGTCCCCCAGGATAAAGGTAATTCGACACTGCGTATGCGGATATATAGCATGT----ATATGTATG------------------------------------------------GCAGAGGATTCCTGCAAACTAATACACTTAAAAATAAATCTGAAAGAT-----------------------------------------------------------------------------------------------AATTTC-------------------------------------CACATTATAGTGTCAACCCCAAGTAATTTGACACACATTCCGCTCGAAGTTTCTCCAA----AC--TATAGTGTCTAA-GATTTGACTTCTTAAATTACTAACCCATATCTTCT-------AC----ATGTGTAAATCAAA-----------A--AAGAACACCAATGTTTACACTCTA----AAAAATCTTTATTGTGCAGATCCAGAAGTATTCGGGCGACGA---TGACGGCAGCTTGGGCAATGCCAACGA | |
| droYak3 | 3R:9140279-9140642 + | TGTACACCTCTGCGGACGAGA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGATCAGGATGAGGCTCCCAGGATAAAGGTAATTCGATACCCATTATGCGGATGCAT-----------ATATGTATT------------------------------------------------GCGGAAGATTGCCTTCAAGAAATACACTTCAAATAAAATCCCACAGAT-----------------------------------------------------------------------------------------------ACTTTC-------------------------------------CACATTATAATTTCAACACCAAGTAATTTGACACACATTCAGTTCGGACTTTCTCCAAAA--AC--TATAATGCCTATTGACTATACTTCTGAAATTACAAAACCATATCTTCT-------AC----ATATGTAAATCAAA-----------ATGGGGATCACCAATGTTTACACTTTT----AAATATCTTTACTGTGCAGATCCAAAAGATTTCGGGCGACGA---TGATGGCGGCTTGGGCAATGCCAACGA | |
| droEug1 | scf7180000407340:15617-15924 - | TATATCACAAAGTCGAGAAGAAAGATCCGGACGATATT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGCCCAAGGATAAAGGTAAATATATGTTTAACATTTGGATACAACAAATGCATGA-------CGGTAAAAACATCCCACAATAATACGTTTCTATATAAACAATGTATTTTTGT--------TAAGTT-------------------------------------------------------TATTTGAGAGTTTTG----ATTT---CT-------------AATCAAATTTTTCCAAAGATCTTGTAGTAGT------TATAAAGAAAAACTGTCTTAAATAATGTTACCTTATATATGT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATGTACATAGATACAGAAAATTTCGGGCGCCGACGATGATGGAGGCTTGGACAATCCCAACGA | |
| droBia1 | scf7180000302402:7816195-7816503 + | TATTGCAATCCGCCGAGACAA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGAACAGGACGAGGGCCCCAGAATCAAGGTAAC--ATTGTTTATAAACAACATCCATACCATATACATATGTATTTA-----------------------------------------------TGT--------TCAATGTACA-TCGTACATACTTACACATGCTAAAGATCTACACTGTATGTATTTTTGGCTACAGTGAGAAATTGAAGGGACTTCCTCTATGCCAATAATATTATTTTATTATTCCAAAG-------------------TATAAGTAAATAATAACAAAAA---TGTATACCCTCC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AA----AACCTTTTTTATAATGCAGATCCAGAAAATTTCGACCGAAGA---AGATG---------------AAACCGA | |
| droTak1 | scf7180000415364:38537-38626 - | TTAAT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TATAACAAAAAAATAGCTAAAA---TGGGAACTTTTC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GA---------AATTTATTTTTTAGATCCAGAAAATTTCAGACGACGA---CGATG---------------ACACCGA | |
| droEle1 | scf7180000491194:240812-241180 + | TATTCCGCACCGATGTGGAGAAAGATCAGGATGAAGATGGTCCACGGATCAAGGTAATATTCTCTATAAAGTTTTGGAGTA------------ACCACAATCAGTGCTGGTGTGATGGGAACTGTAG--GGTCGTTTTGAATAGTAGAT-AATTCTAAAAGATACATTATATACTATGATCTATTAAAATGGTTGTTATAAGGTTGTTCCGACATCTA------------CAACAAATTTTCTCTATTCTCCAATGTCGCCTTCACATATATATAATATATTTATAGTTTTACATATATATAT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATGTGTTTAATAA---------------CTTTCT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AA----AAATGAATATATATTTTAGATCCAGAAAATATCGAGCGAAGACGAAGACGGCGGCTTGGACAACGCTACCGA | |
| droRho1 | scf7180000779488:299238-299572 + | TATTCCGCTCCGATGTGGAGAAAGCTAAAGATGAAGATGGTCCTCGGATCAAGGTAA--CTGTCTATAAGATTCTGGAGCACAGCAAAGTGTTACCCAAAGCAGTGT---TGTGCTGGGAACAGCAGGGGGTCATTTGGAAAAATAGGTGAAATCTAAAAGACACATTCAATCTTATGATCGATTTGAATGTTTGATATAAAGGTGCTGAAACATCTGTAATGTGAGGCACTACAAA------------------------------------------------AGGTTTACAAATATTTATACT------------------------------------------------------------------------------------------------------------AATAGAT--------------------------------------------------------------------------------------------------------------------------------------------------------------------GGT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTCTTTCCTTATATTGTAGATTCAAAAAATATCAAGTGACGAAGGTGATGGCGTCTTGGATAATGCTACCGA | |
| droFic1 | scf7180000454106:2092424-2092484 - | T--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TATGTAGATCCAGAAAATTTCGGGCGACGAAGATGACGGCGGCTTGGAAAATGCTGGCGA | |
| droKik1 | scf7180000301923:188082-188152 - | AATATAAATTCCTTGT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTTAGATCCAGAAAATATCGGCTGAAGA---TGAAGACCAATTGGCCAGTGACAACGG | |
| droAna3 | scaffold_13340:14458326-14458389 - | TATTTA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTCATAGATTCAAAAAATTTCGAGCGCCGA---AGATGTCGATTTCGACAATGCCCCTGA | |
| droBip1 | scf7180000395971:712117-712197 + | CACCCA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAA-----------------------------------------------------------------------------------------------ACTTTC-------------------------------------CATAT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCGTACATAGATTCAAAAAATTTCGAGCGCCGA---AGATGACGATTACGACAATGCCACTGA | |
| dp5 | 2:61650-61710 + | TTT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTAGATCCATAAGATTACGCACGAGGACGCGGATGGCGGCATCGATCATGGGAACGA | |
| droPer2 | scaffold_7:2853442-2853502 + | TTT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTAGATCCATAAGATTACGCACGAGGACGCGGATGGCGGCATCGATCATGGGAACGA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/18/2015 at 09:48 PM