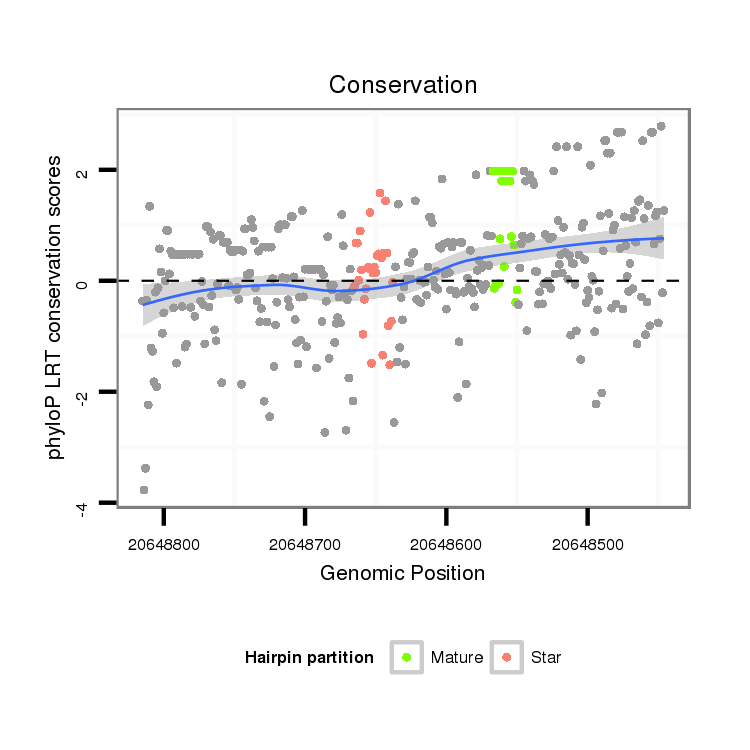
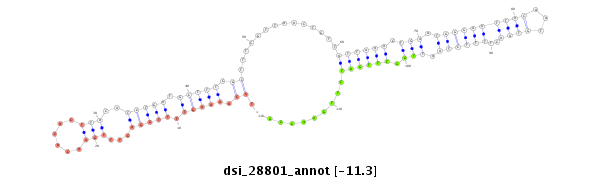
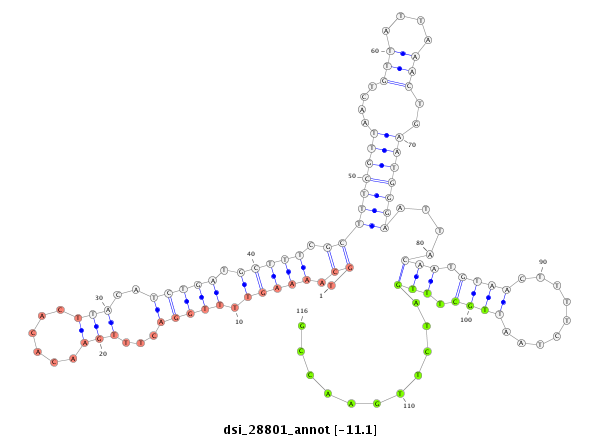
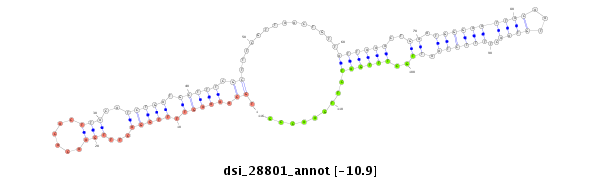
| droSim2 |
3r:20648446-20648815 - |
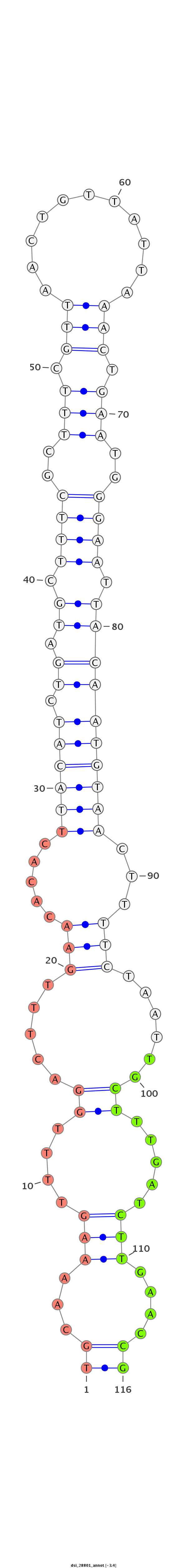
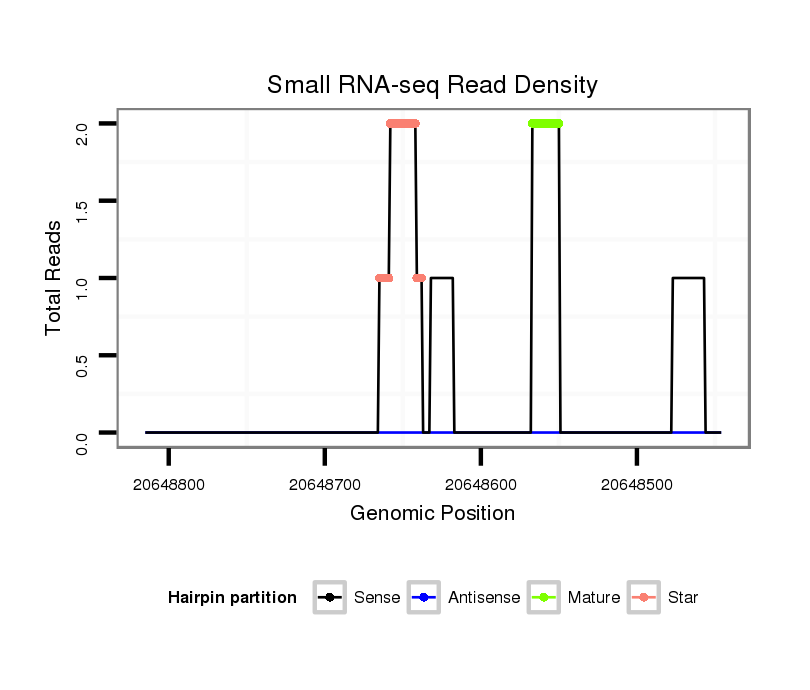
dsi_28801 |
TACAT----------------------------------TACTAGTA---------------------CTAAAA----TTATATAAATA------------------ATAAATTCAAATACATTCCCGTATTT------------------------G----------------------ATAAGTGCATTTTTG------AA----------------------------------------------------------AAAA----AAA-------------------------------------------------------------TAGCAAACTGAAATACTAAATGCTTA--GCCATAAAATTACTATGAAAA---------TG-TAAACT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGCAAA---------------------------------CCAGAA----A---------------------------------------------------------------------------------------------------------------------------------------TGCAA---AAGTTTTGGACTTTGAACAC-------ACTTACA--TC-----------------------------TGA--TGCTTTCGCTTTCG----TTAACTG--TTATTA-AACTGAATGGGAAT-TACAATGTAACTTTTCTAATTGCTTTGATCTTGAACCGCTGTTCGTTTCCGCCGCG--AAA-GTCAATTAATTGATCTAC-CGTTTTCAGTAT------C-------------------------AGCATCACAATGTCGCCAAACAACA---GCGCGGAAAAGCCCGTTAATCCCAACG |
| droSec2 |
scaffold_33:392548-392879 - |
|
TGCATA--------------------------------------GTACGAA---------------------AA----TTATAAAAATA------------------ATAAATTCAAATATATT---------------------------------------------------------------------CG------------------------------------------------------------------AAAA----AAA-------------------------------------------------------------TAGCAGACTGAAATACTAAATGCTTA--GCCATAAAATTACTATTAAAA---------TG-TAAACT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGCAAA---------------------------------CG--AA----A---------------------------------------------------------------------------------------------------------------------------------------TGCAA---AAGTTTTGGACTTTGAACAC-------AC--------------------------------------TGA--TGCTTTCGCTGTCG----TTAACTG--TTATTA-AACTGAATGGGAAT-TACAATGTAACTTTTCTAATTGCTTTGATCTTGAACCGATGTTCGTTTCCGCCGCG--AAA-GTCAATTAATTGATCCAC-CGTTTTCAGCAT------C-------------------------AGCATCAAAATGTCGCCAAACAACA---GCGCGGAAAAGCCCGTTAATCCCAACG |
| dm3 |
chr3R:21133582-21133950 - |
|
TGCAT----------------------------------TCTTTGAA---------------------CAAAAA----TT---------TACAAAATTTACGAAATT-----------TACATTCCGGTATTT------------------------G----------------------ATAAGTGCATTTTCG------------------------------------------------------------------AACA----AAA-------------------------------------------------------------TATCAGACTAATGTGCTAAATACTTA--GCCATAAAAGTACTATTAAAAAACTATTACTA-AAAACT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGCAAA---------------------------------CT--------G---------------------------------------------------------------------------------------------------------------------------------------TGCAA---AAA--TTGGACTTTGAACAC-------ACTAACA--CG-----------------------------TGA--TGCTTTCGCTTTCG----ATAACTG--TTATTA-AAATGAATGGGAAT-CACAATGTAACTTTTTTAATTGCTTTGATCTTGAACGGCTGTTCATTTCCTCCACG--AAA-GTCAATTAATTGATCTAC-CGTTTTCATCAT------C-------------------------AGCATCAAAATGTCGCCAAACAACA---GCGCGGAAAAGCCAGTCAATCCCAACG |
| droEre2 |
scaffold_4820:6909907-6910178 + |
|
AAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TACTAAATATTTAA-GCTGTGAT------------T---------TC-TAAGCTGTGCC----ACGCC------------------------------------------------------------------------------------------------------------------------------------------------------------------------A---------------------------------CTGCAA----A---------------------------------------------------------------------------------------------------------------------------------------TGCAA---ATATTTTAGACTTTGATCAC-------CCTGAAA--AC-----------------------------GGA--GGCTTTCGCTTTCG----TTAACTG--TAATTA-AAATGAATGGAAAA-TGCGATGGAACTTTCTCAATTGCTTTGATCTTGAACCGCTGATCGCTTCCGCCGCG--AAA-CTCAATTAATTGATCTAC-CGCTTACAGCAT------C-------------------------AGCATCGAAATGTCGCCGAACAACA---GCTCGGAAAAGCCCCTCAATCCCAACG |
| droYak3 |
3R:25355342-25355609 + |
|
AATAA----------------------------------TC---AAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATAATCAAAAACTAAATACTTAA-GCTGTGAT------------A---------TT-TATGCT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGCAAA---------------------------------CCAAAA----G---------------------------------------------------------------------------------------------------------------------------------------TGCAA---ATATTTTAGACTTTGCTCAG-------ACA----------------------------------------------TACACTTTCG----TTAACTG--TAATTA-AAATGAATGAGAAA-TGCAACGTAACTATTTTGATTGCTTTGAACTTGAACCGCTGATCATTTCCGCCGCG--AAA-GTCAATTAATTGATCTAC-CGTTTTCAGCAT------C-------------------------AGCCTCGAAATGTCGCCAAACA------GCTCGGAAAAGCCCGTCAATCCCAATG |
| droEug1 |
scf7180000409692:245392-245836 + |
|
AGAAAAA-------------------------------------TTACGGAAATAAATTGTC------CTGAAAAAATT----------------------------C-----------TCTTTCC-GGGTT---------------------------------------------------------------------------------------------------------AATATAACTA-TTATTAAATAATAATAAAAAT------------------------------------------------------------------------------------------A-GTTGTGAA------------A---------TA-GACGTTGAATT----CTGGAACTGGCATAAATATCAATATAAAAACT---------------------------------------------ACTTAA------------------------------------------------TT---GT-------CATTGAAATGAAAATGTCTTCTAAACAA---------------------------------GACAAAA----G-------------------------------------------------------------------------------------------------CGAAAA-----GAAAGTAAACCG---------CTATT---------GAGTTTTAGACTTTGATCAC-------CCAACAA--CCTGTTCAA-TATAACGTTTTTTACATAACATAG--TGCTTTCTCATTCG----TTAACTG--CAATTA-AGATGATTTAAAAA-TTCAATGAAA-TTGTTTAATTACTGTGATCTTGAACTGCTGATCGTTTCCGCTGAGA-----GTCAACTAATTGATCTCC-CGATTACAGATTCAGC------------------------------ACCAACAATGTCGCCGAATACCA---AGTCGGAAAAGCCTGTCAATCCAAACG |
| droBia1 |
scf7180000302136:1952048-1952519 - |
|
TCGGTCGCAAG-----------T---------------------CTACGAATATGTATTGTCTTCTAGCTGAAA----A----------------------------TCAATTTAAAACAAGTTTCCGTCCTTAAAAAAATAATACAAATAAAATATTAAATTAATA------------------------------------------------------------------------------------------------------A-CCGCTAGTTCTTTAAAGTTTTATATCAGCGTTGCTTAACAAATAAAATAAAATAAATA-------------------------------------------------------------------------------------------------------------------------------------------------------------AATGAA------------------------------------------------AAAT-TGCATCCGTT--------GAATATATTATCTGAGCGA---------------------------------GAAAGAA----C-------------------------------------------------------------------------------------------------CGAAAA-----GAAAGAAAGTTG-----TTCCCTGTT---------ATATTTTCCAGTTTGTTCAA-------CAATACA--CCTGTTGAATTTTAAAGATTTTCACGTTATACGG--TGCTTTCTCTGCCG----TT----C--TAATTA-AGATGAATGGGTG--TATAATAAAA-ATGTTTAATTACTTTGATCTTGAACCGCTGATCGTTGCCGCCGAGA-----GTAAACTAATTGATTTCC-CGATTACAGCAT------C-------------------------TGCACCAAAATGTCGCCGAACAGCA---ACTCTGAGAAACCAGTCAATCCCAACG |
| droTak1 |
scf7180000413457:81583-82046 + |
|
TAAAT---------------------------------------CTACGGAAATACCTTT--------------------AAATAAACA------------------AA----------------------------------------------------------------------------------------------------TGATTT------------AAAGCTGGTGGAGCAAGTGA-TTTTTAAATA-CATTAAAAAT------------------------------------------------------------------------------------------A-TAAAATAA------------T---------TA-TATGTCGCTTACAAACATGAGTTTCAGTGAATTTCAAACAGAAAATT---------------------------------------------ATTAAA------------------------------------------------AA---TTCAAC---CATTGTAATG--------ATCTGTGTAAAGAGAAGCGTATAAAAAATGTAAATCTAAATAAGACAAAG----C-------------------------------------------------------------------------------------------------GGAAAA-----GAAAGAAAGTTT-----T-------T---------ATATTTTCGTCTTTGGTCAC-------CCAT----------C-AC-TTTTAAGATTTTCACGTAATAGGC--AGCTTTCTCTGTCG----TAAACAC--TAATTA-AAACGAACGGGAAT-TGTAATGAAA-TTGTTTAACTATTTTGATCTTGAACCGCTGATCATTTCCGTCGAGA-----GTCAATTAATTGATTTCCACGATTACAGCAT------C-------------------------AGAACAAAAATGTCGCCGAACAGCA---ACTCGGAAAAGCCAATCAATCCAAACG |
| droEle1 |
scf7180000490967:21520-21914 - |
|
TATAT----------------------------------TATCTCTA---------------------CTGAAA----T----------------------------C------------CATTTCCGAGTTT------------------------GTTTTC---------------------A---------------------------------------------------------------------------------------------------------TTGTATTTAGGGGTTTG------------------------------------------------------------------------------------------------------------------TTAATATGACTCAAAAAAGCGCTTTATATTAAAGCTTGAAACAACAGTGTGTGAAATACAATAGCATTAAA------------------------------------------------GAGT-CTCAACTGCT--------AAAAGTGGTATTTAAGTTA---------------------------------G-----------------------------------------------------------------------------------------------------------AGGAAAACTA-----------------------------AGCGAATAACATTTTGGACCTTGATCTA-------C---------CTGTT-CC-TATTGCGATTT-TACGTAATA-----TGCTTTCTATTTCG----TTAACTG--TAATTA-AAATGAA----AAA-AACGAA--------GTTAATTGCTTTGATCTTGAACCGCTGATC---TCCGCCGAGA-----GTGAATTAATTGATCTAC-CGATTACAGCAA------C-------------------------AGCAGCAGAATGTCACCGAACAACG---ACTCGGAAAAGCCACTCAATCCCAACG |
| droRho1 |
scf7180000779739:29471-29937 + |
|
TAAACTTATCAA---------------------------TCAGTTGA---------------------CAACAA----TT---------TATAA----------------------------------------AAACAATAATAAGAACACAATATATTTTAAGGAAACGAACTCACCCATAAA---------------------------------------------------------------------------------------------------------TTGTATCAAAGGTTTTGAATGTAAAAGATGTGATAAGAAAAGGAAGAATAATTAACAACTAAATATCTAAGGCCATTAA--------------------------------------------------------------------AATT---------------------------------------------ATTAAA------------------------------------------------AGGT-TTCAAC---T--------AAAAGTGTTATCTTGATTA---------------------------------A-----------------------------------------------------------------------------------------------------------AGAAAAA-----GTAAATAAATTTGA-------ATA-TAAGCAA------ATTTGGACCTTGCTCAT-------TTAAATA--CTTGAC-AC-TATTACGATTTGTACGTAATA-----TGCATTCTATTTAG----ATAACTG--TTATTA-AGATTAATGGAAAA--TCAATTAAA-GTTTTTAATTGATTTGATCTTGAAGTGCTGATCGTTTCCGCCGATA-----GTGAATTAATTGATCAAA-CAATTACAGCAT------C-------------------------AGCGCCATAATGTCTCCGAACAACA---ACTCGGAGAAGCCATTCAATCCCAACG |
| droFic1 |
scf7180000454055:1048588-1049077 - |
|
TTGCTAGACAT-----------T---------------------TTATGGATATAA---------------------TT----------------------------C-----------ACTTTCC------T------------------------C----------------------ATAAATGCAATCTGGTTTATAAAAACAAATGAACTAAATGAA---CTCAGACTTTTTAAACAAGTGA-TTT--------------------------------------------------------------------------------------------------------TCTAGTG-------------------------------------------------------------AATCGCT-----TTAATT---------------------------------------------ATTATAAAGGTTTACCTACTTTTGAGTTTAAAGACAACAACACCTACAAGTTAAAGATATTAAACCC-C--------TAATCTGTTATTTTAACCA---------------------------------A-----G-----------------------------------------------------------------------------------------------------GGGAAAACTAATGAAAGAAAGTTT-----TTGCATAATA---AA---A--AAATCGACTTTGATCAC-------TAATACACATCTGTTAAC-TATTGAGATTTTCACGTAACATAT--TGCTTTCTCTTTCG----ATAACCGACTAATTA-CAATGAATGTGAAA-TTCAATGAAA-TTGTTTAATTGCTTTGATCTTGAACCATTGATCATTTCCGCCGAGT-----GTAAATTAATTGATCTTC-CGATTGCAGCAA------C-------------------------GAATCCAGAATGTCGCCGAACAGCA---AATCGGAAAAACCAGTCAATCCCAATG |
| droKik1 |
scf7180000302355:1386009-1386141 + |
|
TGT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAA-TTGTTTAATTGCTTTGATCTTGGACTGTTGATCGTTTCCGCTGTCT-----GTCAATTAATAGATTTTTCTGATTGCAG---------C-------------------------AGCCTCAGAATGTCGCCGAACAGCA---GCTCGGAAAAGCCAGTCAATCCGAACG |
| droAna3 |
scaffold_13340:1331763-1332178 - |
|
TTAATGAAAAATTCAAAGCGAATATGTGAATAGAAATACTATGTGTG----------------------------------------------------------------------------------------------------------------------------------------------AG------CTTAAACAAATTAAAACTAACTGAAGAAATCA-------------ACAAAGCTTTC------CGATAA----A-AAGACAAATGTTGGAAGTC--------------------------------------------------------------------------------------------------------------T-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T------------------TTGTCAGAATGACACGGCAACTTCCCATTGTAGAATATAGTATCTACGCATTGAGAAAA-------------------AC-----TGTATTC------------------------TATAGTCACACCCAATGTAGACA-----------T-TTTAAGGTTTTC---CGATTTGCAATTCACTCGATTTAAACTGT----TC--TGGGTA-AAAATAAGATAAATGCGCATGGAAG-TTGTGTAATTACTCTGAACTTGAACCTCTAATCATTTCCTCATGTG-----CTCAATTAATTTCTATAT-TAGTTACAGTGC------C-------------------------AG--TGAAAATGTCGCCAAACAACAAAAGCGAAGATAAACCAGTGAACCCCAACG |
| droBip1 |
scf7180000396712:493157-493558 + |
|
AATAC----------------------------------TATTTGTG---------------------T--------------------------------------------------------------------------------------------------------------------------------CATAAAAAAATTAAAACTTAGTGGGGAAATCA-------------AGAAAGCTTTC------CGATAA----AAAAGACAAATGTTGAAACTC--------------------------------------------------------------------------------------------------------------G-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTTGTCAGAATGACACGGCAACTTCCCATTCAAGAATATAGTATCAACGCATTGAGAAAACGAATA-----CTAAATAAACCTACATGTTTCC------------------------TATAGTCACACCCAATGT---TA-----------C-GTTAAGGTTTTC---CAATATGCAATTTACTCGTCACACAGTGT----TC--TGGGTAGAAATTAAAGCGAAA-TGAATTGTAA-TTTTCTAATTGCTACGAACTTGAACCTCCAATCATTTC-GCCATGTGTATGCCCAATTAATTTGTATAT-TAATTTTAGTTC------C-------------------------AGT--GATTATGTCACCAAACAACAAAAGCGAAGAGAAACCTGTAAACCCCAACG |
| dp5 |
2:20537373-20537721 + |
|
AGTGA----------------------------------TACTCTTAAGAATAT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGTGCTATATATGTAC--TTTCGAC------------T---------GCTT-----GCGCC----ATTCC-----------------------------------TAAATGAATGAAATTATAGTAAATTATACGTAGCGTAACTGAA------------------------------------------------CGAT-AAAGCCCGT----------------------------------------------TGTAGACTTAGATAAGTCAATAGGAAACGAAACAAGTTAGACAGTGTCTGAAAGTGTAATTTGGCCTTTCACCATGACGT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AACAATTT-CTATACATAGAAAA-TGCTTTAGAG-CCGTTTTATTGTTTTGATCTTGAACCCACGATCGTTTCCGAT---------CTCTAGTCTCTGATCTCT-TACTTACAG--T------C-------------------------AGTCCCAATATGTCGCCAAACAGCA---GCTCAGAGAAGCCGGTGAATCCCAATG |
| droPer2 |
scaffold_3:3291568-3291891 + |
|
TATATA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTAC--TTGTGAC------------T---------GCTT-----GCGCC----ATACC-----------------------------------CAAATGAATGAAATTATAGTAAATTATACGTAGCGTAACTGAA------------------------------------------------CGAT-AAAGGCCGT----------------------------------------------TCTAGACTTAGATAAGTCCATAGGAAACGAAACAAGTTAGACAGTGTCTGAAAGTGTAATTTGGCCTCTCACCATGACGT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AACAATTT-CTATACATAAAAAA-TGCTTTAGAG-CCGTTTTATTGTTTTGATCTTGAACCCACGATCGTTTCCGAT---------CTCTAGTCTCTGATCTCT-TACTTACAG--T------C-------------------------AGTCCCAATATGTCGCCAAACAGCA---GCTCAGAGAAGCCGGTGAATCCCAATG |
| droWil2 |
scf2_1100000004943:13491240-13491287 + |
|
T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAAAATGTCACCTAATACTA---AGTCAGAAAAGCCTCAAAATCCAAATG |
| droVir3 |
scaffold_13047:2273819-2273904 + |
|
AGGCCAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTACCAGATCTTT-CAGTGCCCACTTGCATATCAGTCAGAATGCCCGATAACAGCAACAAAT--------------------------TCTGTGAATCGA-------ATGATG |
| droMoj3 |
scaffold_6540:22635328-22635408 - |
|
AGGCGAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTATTGGATC----TGTTCTCAGTTG------C-------------------------TCAGTCAGAATGCCCGATAACAGCA---ACA-AGTTCTGCGAGTCAAATCATACA |
| droGri2 |
scaffold_14906:9123293-9123329 + |
|
AAT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AACAGCG---ATTCTAGTAAACCTGTGAATCCCAACG |