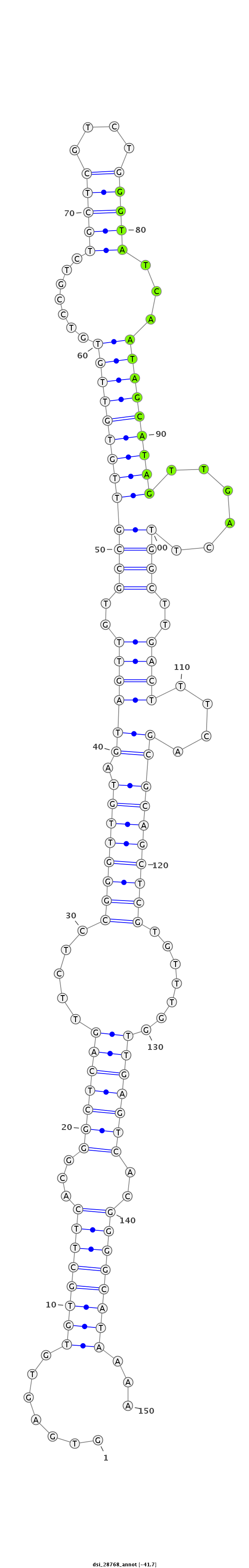
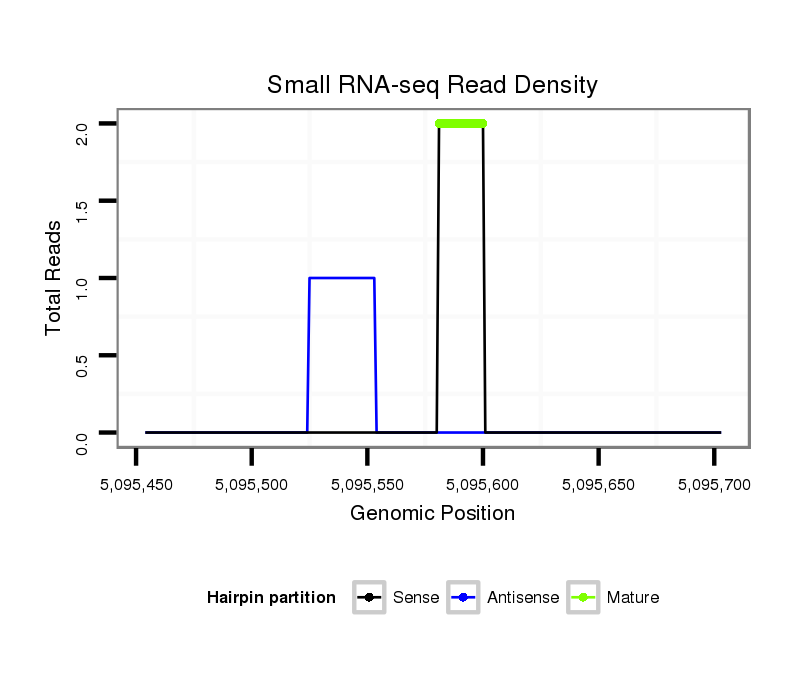
ID:dsi_28768 |
Coordinate:2r:5095504-5095653 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -41.5 | -41.3 | -41.2 |
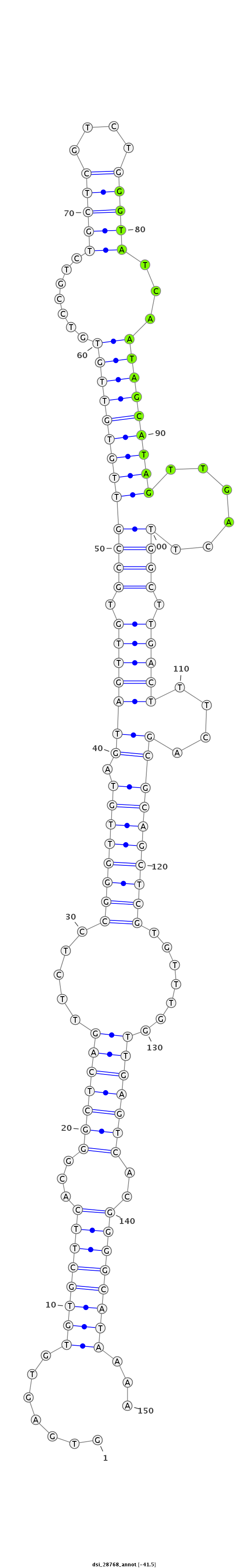 |
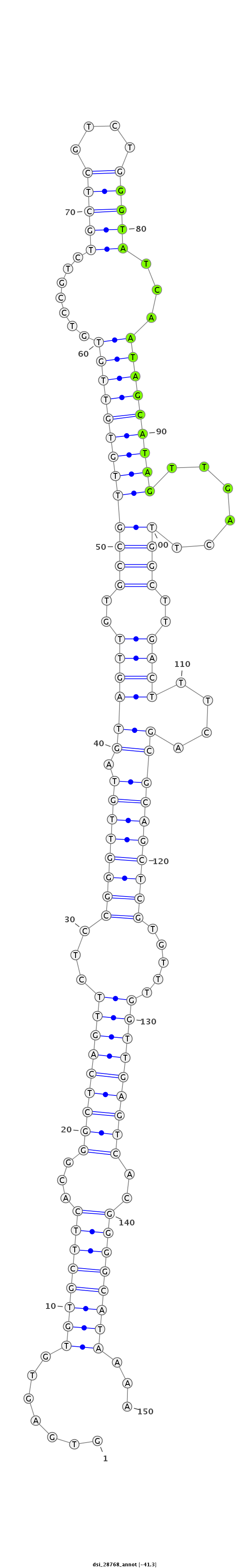 |
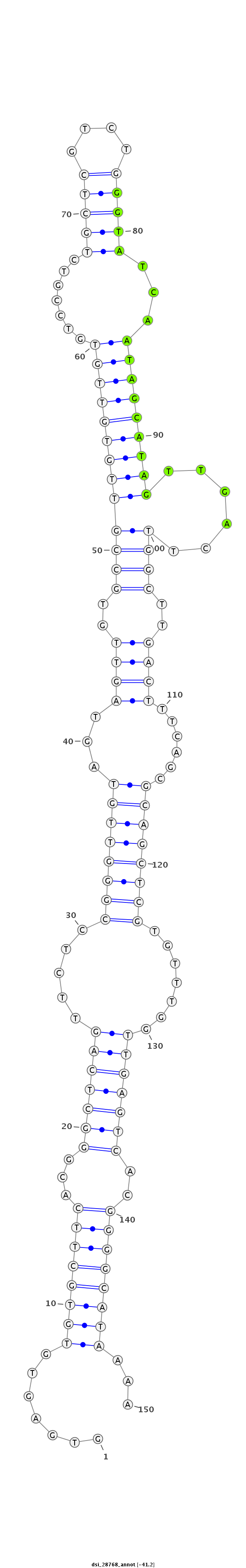 |
intergenic
No Repeatable elements found
|
GCTGGTGCAGGGCGTCCAGGGCGCCCAGCTGCTCATACCCACAGCCCAAGGTGAGTGTGTGCTTCACGGGCTCAGTTCTCCGGGTTGTAGTAGTTGTGCCGTTGTGTTGTGTCCGTCTGCTCGTCTGGGTATCAATAGCATAGTTGACTTGGCTTGACTTTCAGCGCAGCTCGTGTTTGGTTGAGTCACGGGGCATAAAAAGCTCGTAAAGCGATTAGGGCAACTTTCAATTTCCTCCTCGCGTGGGGCG
**************************************************.......((((((((...(((((((.....((((((((.((((((..(((((((((((((.......(((((....)))))...)))))))))......))))..))))....)))))))))).......)))))))..))))))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | O001 Testis |
SRR553487 NRT_0-2 hours eggs |
SRR902009 testis |
SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................GGTATCAATAGCATAGTTGA....................................................................................................... | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................TGAAGTAGTTGTGGCGTTTT................................................................................................................................................. | 20 | 3 | 18 | 0.28 | 5 | 0 | 0 | 5 | 0 | 0 | 0 |
| ........................................................................................................................................AGCATCGTTGACTTG................................................................................................... | 15 | 1 | 11 | 0.27 | 3 | 0 | 0 | 0 | 3 | 0 | 0 |
| .............................................................................................................TGTCAATCTGCTCGTTTGGGT........................................................................................................................ | 21 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................AAAGCTCGTACAGCGGTT.................................. | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................TCCGGGGTGTCGTAGTT........................................................................................................................................................... | 17 | 2 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................CCGGGTGGTAGTAGC............................................................................................................................................................ | 15 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CGACCACGTCCCGCAGGTCCCGCGGGTCGACGAGTATGGGTGTCGGGTTCCACTCACACACGAAGTGCCCGAGTCAAGAGGCCCAACATCATCAACACGGCAACACAACACAGGCAGACGAGCAGACCCATAGTTATCGTATCAACTGAACCGAACTGAAAGTCGCGTCGAGCACAAACCAACTCAGTGCCCCGTATTTTTCGAGCATTTCGCTAATCCCGTTGAAAGTTAAAGGAGGAGCGCACCCCGC
**************************************************.......((((((((...(((((((.....((((((((.((((((..(((((((((((((.......(((((....)))))...)))))))))......))))..))))....)))))))))).......)))))))..))))))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
SRR902009 testis |
SRR553485 Chicharo_3 day-old ovaries |
SRR902008 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................AGTCAAGAGGCCCAACATCATCAACACGG...................................................................................................................................................... | 29 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................AGTCAAGGGGCCCAAC................................................................................................................................................................... | 16 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................ACCAACTCAGTGCCCCTTAG..................................................... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................CGAGTCAAGAGGCCCAACAGC................................................................................................................................................................ | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................AGTCAAGGGGCCCAA.................................................................................................................................................................... | 15 | 1 | 6 | 0.50 | 3 | 0 | 0 | 3 | 0 | 0 | 0 |
| ..........................................................................................................................................................................AGCACAAGCAAACTCAATGCC........................................................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................ACCACCTCAGTGCCCCTTA...................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................CAGAACACAGTCAGACGA................................................................................................................................. | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............AGGTCCCGCGGGACG............................................................................................................................................................................................................................. | 15 | 1 | 16 | 0.19 | 3 | 1 | 0 | 1 | 0 | 1 | 0 |
| .......GTCCTGCAGGTCCCACCGGTC.............................................................................................................................................................................................................................. | 21 | 3 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................AGTCAAGTGGCCCAA.................................................................................................................................................................... | 15 | 1 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................AGTCAAGAGGTCCAA.................................................................................................................................................................... | 15 | 1 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............AGGTCCCGCGGGCCG............................................................................................................................................................................................................................. | 15 | 1 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................AAGTCAAGAGGCCCA..................................................................................................................................................................... | 15 | 1 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................AGAGTCAAGGGGCCCAA.................................................................................................................................................................... | 17 | 2 | 16 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............AAGGTCCCGCGGGTC.............................................................................................................................................................................................................................. | 15 | 1 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................ACCCAATCAGTGCCCC......................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................AGTGCCCAAGTCAGGA........................................................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................TCGACACAACACAGGGAGA.................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/23/2015 at 09:32 AM