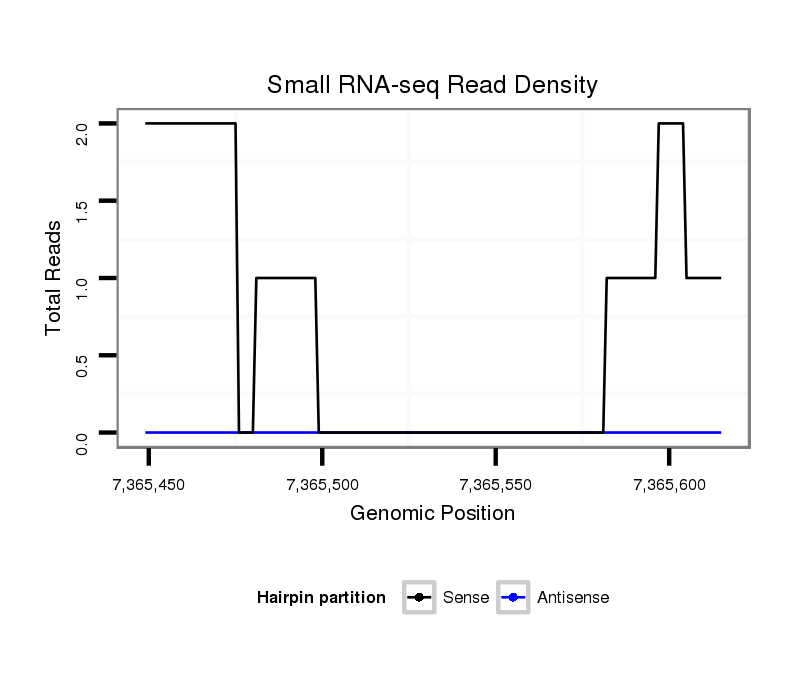
ID:dsi_28705 |
Coordinate:3l:7365499-7365565 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
exon [3l_7364743_7365498_+]; CDS [3l_7364743_7365498_+]; CDS [3l_7365566_7366220_+]; exon [3l_7365566_7366220_+]; intron [3l_7365499_7365565_+]
No Repeatable elements found
| ##################################################-------------------------------------------------------------------################################################## TTGATCGGTACTATGAGATTCTACAGAATGATCCCACTTTCGAGGACAGTGTAAGTTAATAAGCCGAGTTTTCTATCAGCCTATTAGAAAATATAAAACATTGAATTCCTTTTTTAGGACACCGATGGTCGCGAGAACGCACAGCGCATCTTCGATCGCGTCTCGGA **************************************************.....(((((.......((((((((.........)))))))).......))))).............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
M023 head |
M025 embryo |
SRR553487 NRT_0-2 hours eggs |
M053 female body |
SRR902009 testis |
SRR553485 Chicharo_3 day-old ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
GSM343915 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................AATGATCCGACTTTCGTG........................................................................................................................... | 18 | 2 | 6 | 2.17 | 13 | 10 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TTGATCGGTACTATGAGATTCTACAGA............................................................................................................................................ | 27 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................TCTTCGATCGCGTCTCGGA | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TTGATCGGTACTATGAGATTCTCCAGA............................................................................................................................................ | 27 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............AGATTCTACAGGACGATC...................................................................................................................................... | 18 | 2 | 2 | 1.00 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ................................CCCACTTTCGAGGACAGT..................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................GTCTTCGATCGCGTCTCGGA | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGAACGCACAGCGCATCTTCGAT........... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................TCGAGGACAGTGTATG................................................................................................................ | 16 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................CAATGATCCGACTTTCGTG........................................................................................................................... | 19 | 3 | 20 | 0.40 | 8 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................CCAGAATGATCCCACT................................................................................................................................. | 16 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................CGAGGACAGTGTATG................................................................................................................ | 15 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................ATCGAGGACAGTGTA.................................................................................................................. | 15 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................ATCGAGGACAGTGTATG................................................................................................................ | 17 | 2 | 12 | 0.17 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................TCGAGGACAGTGTCA................................................................................................................. | 15 | 1 | 17 | 0.12 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................AATTCCTGCTTTAGGAGACC............................................ | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................................CCATCGAGGACAGTGTA.................................................................................................................. | 17 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................ACAGATTGATCCCTCTTT............................................................................................................................... | 18 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................................GAGGACAGTGGAACTTCAT........................................................................................................... | 19 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................CCCGATCGAGGACAGTGT................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
|
AACTAGCCATGATACTCTAAGATGTCTTACTAGGGTGAAAGCTCCTGTCACATTCAATTATTCGGCTCAAAAGATAGTCGGATAATCTTTTATATTTTGTAACTTAAGGAAAAAATCCTGTGGCTACCAGCGCTCTTGCGTGTCGCGTAGAAGCTAGCGCAGAGCCT
**************************************************.....(((((.......((((((((.........)))))))).......))))).............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
M024 male body |
M025 embryo |
SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................AAAAAATCAAGTGTCTACCAG..................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................AAAAATCAAGTGTCTACCAG..................................... | 20 | 3 | 11 | 0.18 | 2 | 0 | 0 | 2 | 0 | 0 |
| ..................AAAATGTCTTACTAAGGT................................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................AAGCTTCTGTCAGATTC................................................................................................................ | 17 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................TTTCTTACTAGGGTG.................................................................................................................................. | 15 | 1 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 |
| .....................ATGTCTTACTAAGGT................................................................................................................................... | 15 | 1 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................AGGGTGATAGCTCCT......................................................................................................................... | 15 | 1 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................CAAGGGTGATAGCTCCT......................................................................................................................... | 17 | 2 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................TTACTAGGCTGAAAGCACA.......................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................ATGTCTTACTAAGGTGGTA............................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................ACCATGTCTTACTAAGGT................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
Generated: 04/24/2015 at 06:04 AM