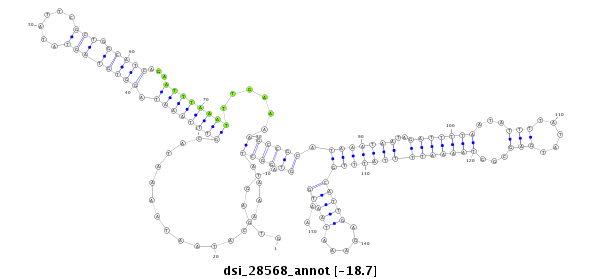
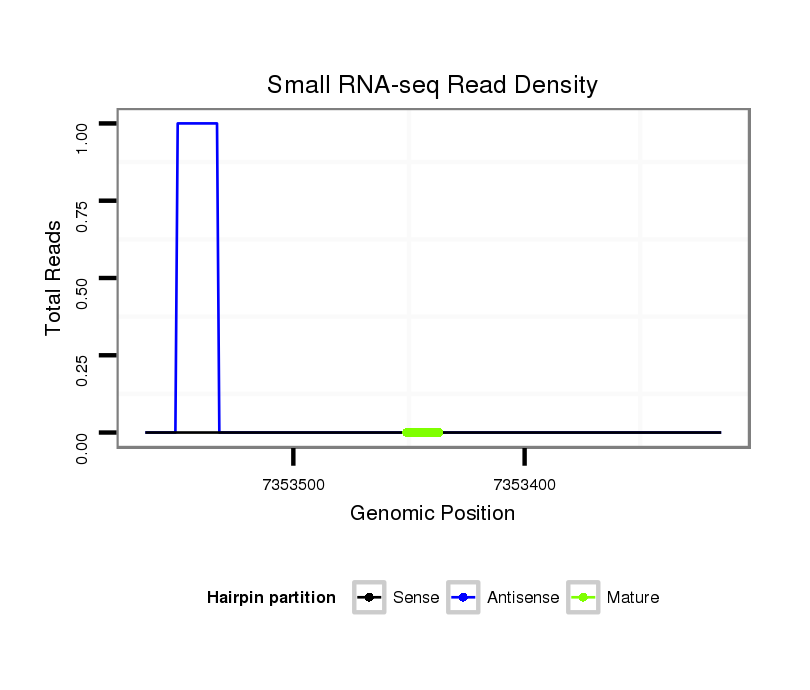
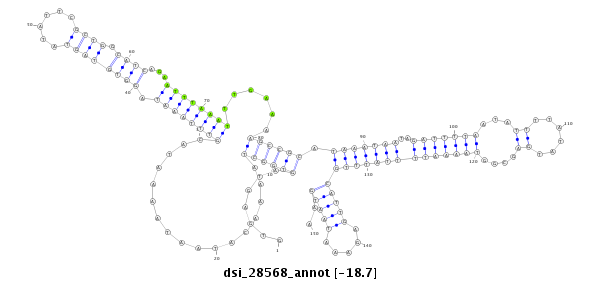
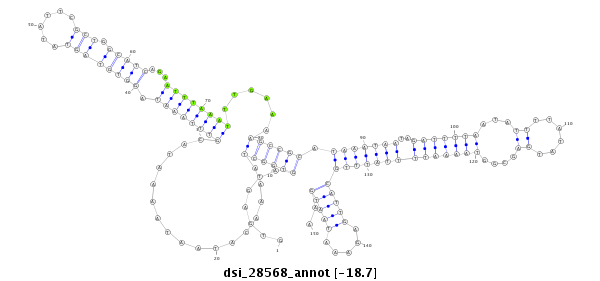
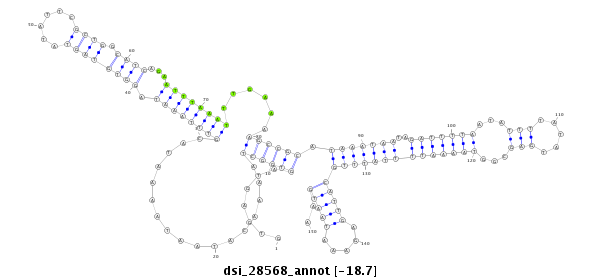
ID:dsi_28568 |
Coordinate:3l:7353365-7353514 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -18.7 | -18.7 | -18.7 |
 |
 |
 |
five_prime_UTR [3l_7353546_7353665_-]; CDS [3l_7353515_7353545_-]; exon [3l_7353515_7353665_-]; intron [3l_7352617_7353514_-]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| mature | star |
| ##################################################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TCCACCGCTCATTGGCGCCATGCAACTCATGCTCCGTTTGAACCCAAAAAGTGAGTAGTGGCTAAAACATAATAAAATACGTTTAAATAGGTGTAGTATATTCGCTGGCATCAGAATTTAAATTTGAAAAGCCGCATAAATAATAGATTTTAATATTTTATATGAGCGGTAAAATTTTATTTGCATTGAGAAATAATGAAATGAATAGAGCTGCAATATCAATTACCTAAAACTGCTATCAATCAAAACG **************************************************.......((((((.................((((((((.((((((((......))).)))))...))))))))......)))))).(((((((..(((((((...(((.....)))...))))))))))))))(((((.....)))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
M053 female body |
SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
M024 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................AACGAATAGAGCTGC.................................... | 15 | 1 | 11 | 0.55 | 6 | 6 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................AGTATATTCGCTGGGATTAGG....................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................CAAACGAATAGAGCTGC.................................... | 17 | 2 | 20 | 0.35 | 7 | 7 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................AAACGAATAGAGCTGCGA.................................. | 18 | 2 | 10 | 0.30 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................TTCAATAGCTGTAGTATATG.................................................................................................................................................... | 20 | 3 | 11 | 0.18 | 2 | 0 | 0 | 0 | 2 | 0 | 0 |
| .........................................................................................................................................................................................................................................TTCTATCAATCAAAAC. | 16 | 1 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................GAATTTAAATTTGAA.......................................................................................................................... | 15 | 0 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................................................................TGAATAGAGCTGCGA.................................. | 15 | 1 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTGAGTCGTGGCTAA......................................................................................................................................................................................... | 15 | 1 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................AATAGGTGTAGAATA...................................................................................................................................................... | 15 | 1 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................AAAAAACGAATAGAGCTGCA................................... | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................ATGGTCCGTTTGAACTGAA........................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
AGGTGGCGAGTAACCGCGGTACGTTGAGTACGAGGCAAACTTGGGTTTTTCACTCATCACCGATTTTGTATTATTTTATGCAAATTTATCCACATCATATAAGCGACCGTAGTCTTAAATTTAAACTTTTCGGCGTATTTATTATCTAAAATTATAAAATATACTCGCCATTTTAAAATAAACGTAACTCTTTATTACTTTACTTATCTCGACGTTATAGTTAATGGATTTTGACGATAGTTAGTTTTGC
**************************************************.......((((((.................((((((((.((((((((......))).)))))...))))))))......)))))).(((((((..(((((((...(((.....)))...))))))))))))))(((((.....)))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | O001 Testis |
M053 female body |
M025 embryo |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|
| ..............CGCGGTACGTTGAGTACG.......................................................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................TTCACATAATATAAGCGA................................................................................................................................................ | 18 | 2 | 6 | 0.33 | 2 | 0 | 2 | 0 | 0 |
| ..........................................................................................................CCCTAGTCTGAATTTTAAA............................................................................................................................. | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 |
| ......................................................................................AGTCCACATAATATAAGCGA................................................................................................................................................ | 20 | 3 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 |
| ............................TACGAGTCTAACTTGTGTTTT......................................................................................................................................................................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 05:51 AM