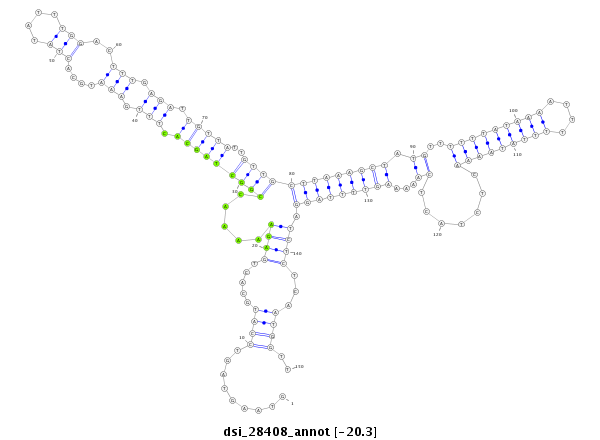
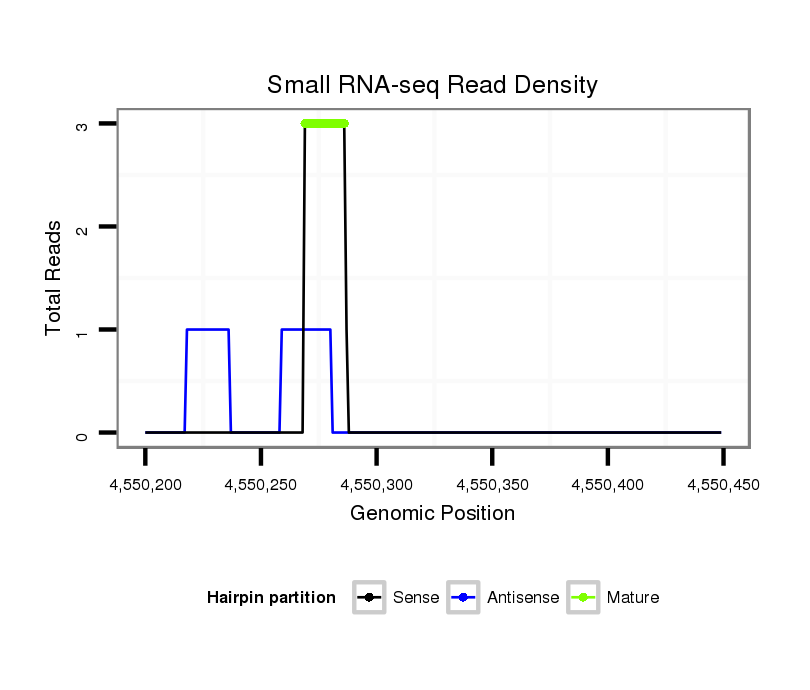

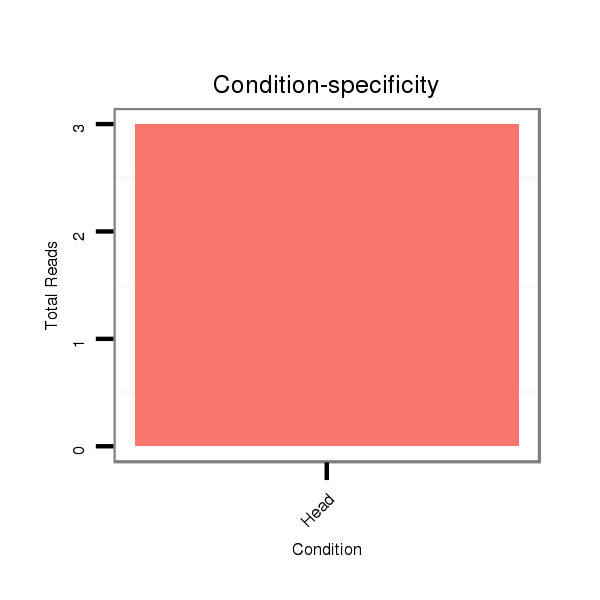
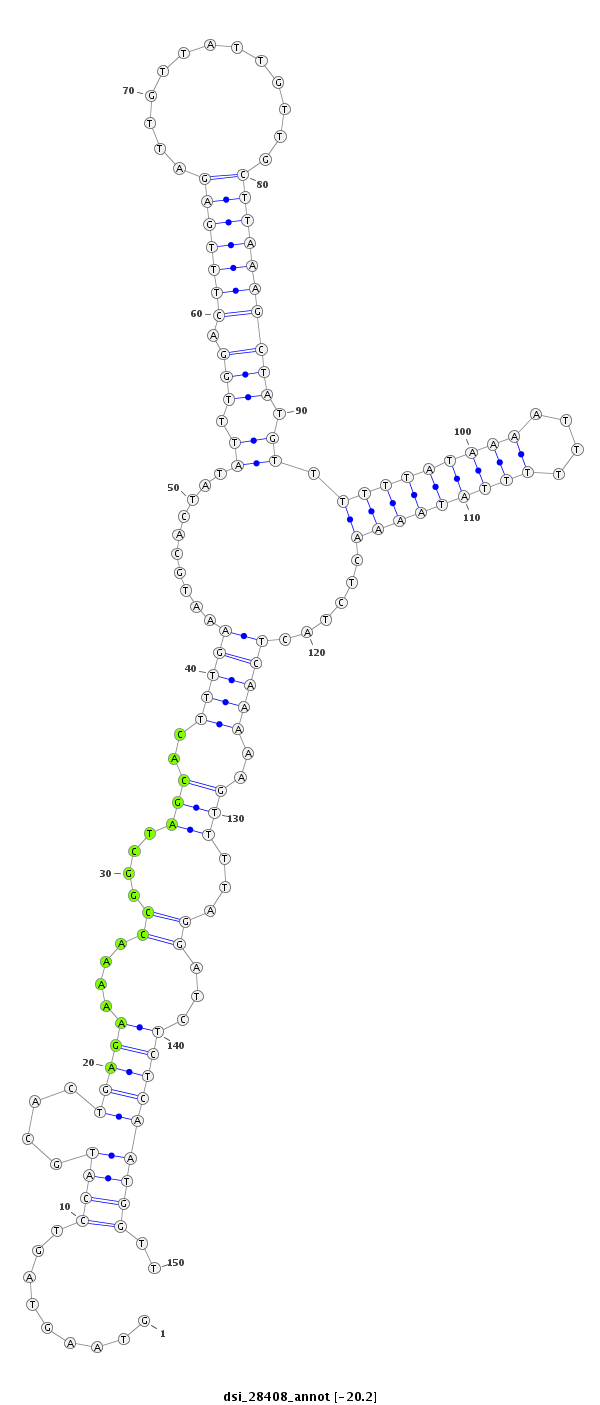
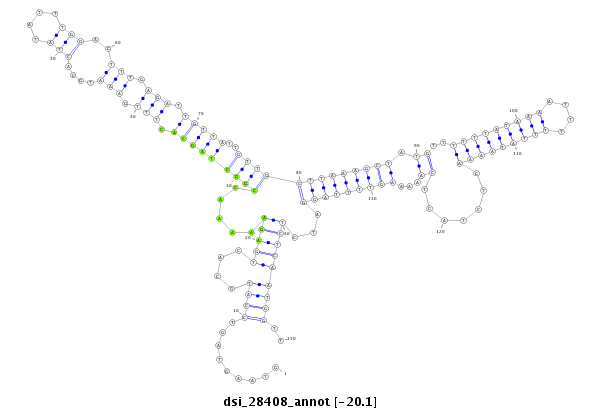
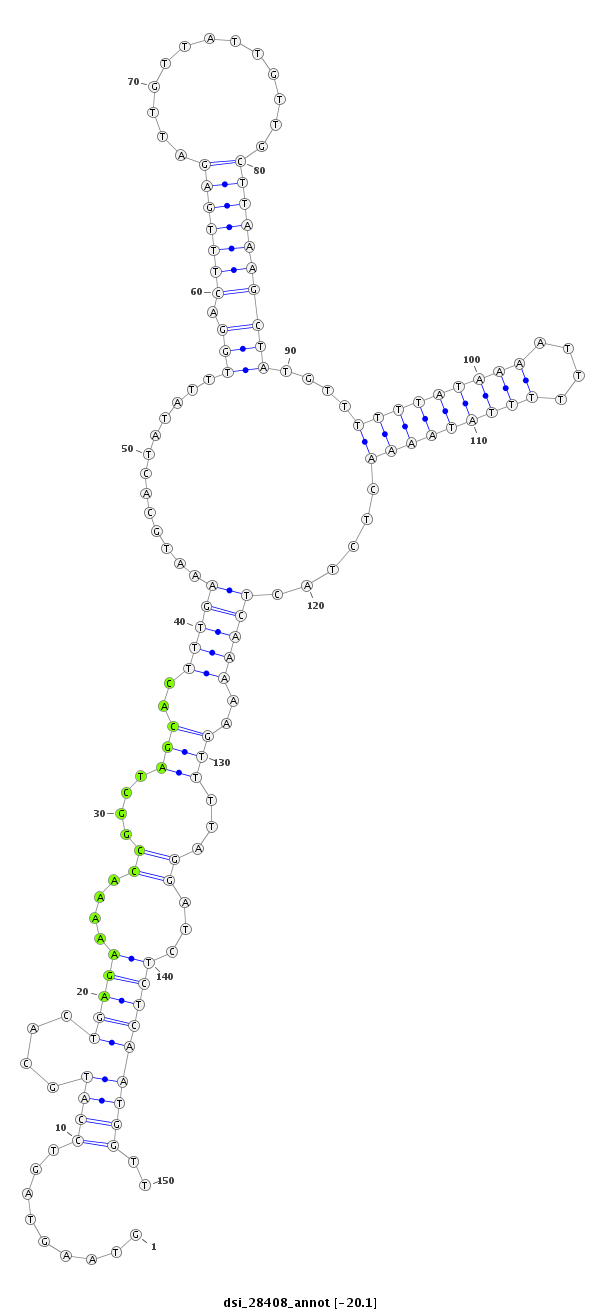
ID:dsi_28408 |
Coordinate:3r:4550250-4550399 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -20.2 | -20.1 | -20.1 |
 |
 |
 |
intergenic
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
|
GCGCTCTAATACCAGTGGCTCCACCACCAGATCCTACGACAGCGGAAGCGGTAAGTAGTCCATGCACTGAGAAAAACCGGCTAGCACTTTGAAATGCACTATATTTGGACTTTGAGATTGTTATTGTTGCTTAAAGCTATGTTTTTTATAAAATTTTTTATAAAACTCTACTCAAAAAGTTTTAGGATCTCTCAATGGTTTACTTAACTTATAAAGTATCTAAAATACATTATTGTGTTGCAATTTAAAG
**************************************************.........((((.....((((.....(((((((((.(((.(((....(((....)))..))).))).)))))..))))(((((((((.((..(((((((((....))))))))).......))...))))))))).))))...))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | O002 Head |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................TTCTAAAACTCTACTCAACA......................................................................... | 20 | 2 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| .....................................................................AGAAAAACCGGCTAGCAC................................................................................................................................................................... | 18 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .....................................................................AGAAAAACCGGCTAGCACT.................................................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................AAAAAGTTTTATGATGTCTGAA....................................................... | 22 | 3 | 6 | 0.50 | 3 | 0 | 3 | 0 | 0 | 0 |
| .........................................................................................................TGGACTGTGAGATTG.................................................................................................................................. | 15 | 1 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 |
| ................................................CGGTAAGCAGTCCATGCTG....................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................GACTTTGAGATTGACAT.............................................................................................................................. | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
|
CGCGAGATTATGGTCACCGAGGTGGTGGTCTAGGATGCTGTCGCCTTCGCCATTCATCAGGTACGTGACTCTTTTTGGCCGATCGTGAAACTTTACGTGATATAAACCTGAAACTCTAACAATAACAACGAATTTCGATACAAAAAATATTTTAAAAAATATTTTGAGATGAGTTTTTCAAAATCCTAGAGAGTTACCAAATGAATTGAATATTTCATAGATTTTATGTAATAACACAACGTTAAATTTC
**************************************************.........((((.....((((.....(((((((((.(((.(((....(((....)))..))).))).)))))..))))(((((((((.((..(((((((((....))))))))).......))...))))))))).))))...))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553485 Chicharo_3 day-old ovaries |
M053 female body |
SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR902008 ovaries |
GSM343915 embryo |
M024 male body |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................GAGGTGGTGGTCTAGGATG..................................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................GGTACGTGACTCTTTTTGGCCG......................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................GAAGCTGTCGCTTTCGCCA...................................................................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................AAAAAATATTTTGCGATAAGATT.......................................................................... | 23 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................AAAAAATATGTTGATAGGAGT............................................................................ | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................TCGCTATCCATCAGGTTCGT........................................................................................................................................................................................ | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................GTGGTGGGCTAGGAT...................................................................................................................................................................................................................... | 15 | 1 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................CAATAACAAGGAATCTCG................................................................................................................. | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................TCAAAGTCCTAGAGA.......................................................... | 15 | 1 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................AGGTACGTGAATCTTT................................................................................................................................................................................ | 16 | 1 | 15 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGTGATTCATCAGGTACGT........................................................................................................................................................................................ | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................CCGGGGTGATGGGCTAGGA....................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................................ATATCTTGAGATGTGTTTTG........................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 09:34 AM