| droSim2 |
3r:6413300-6413549 - |
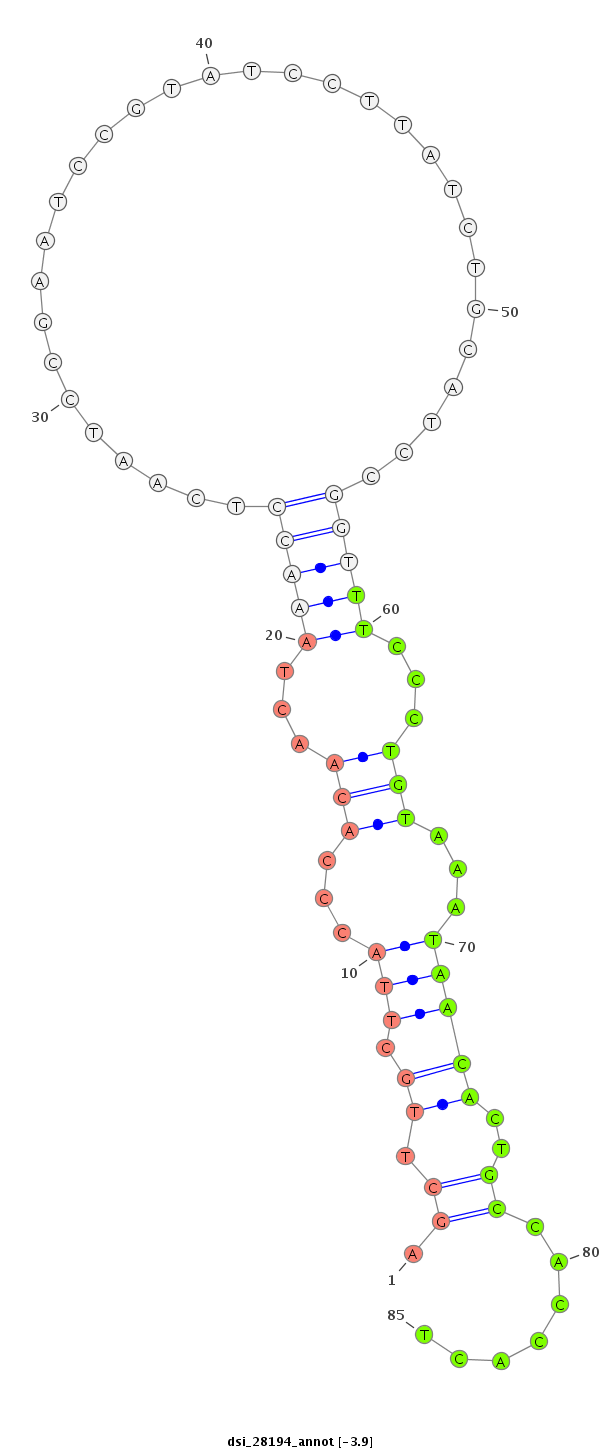
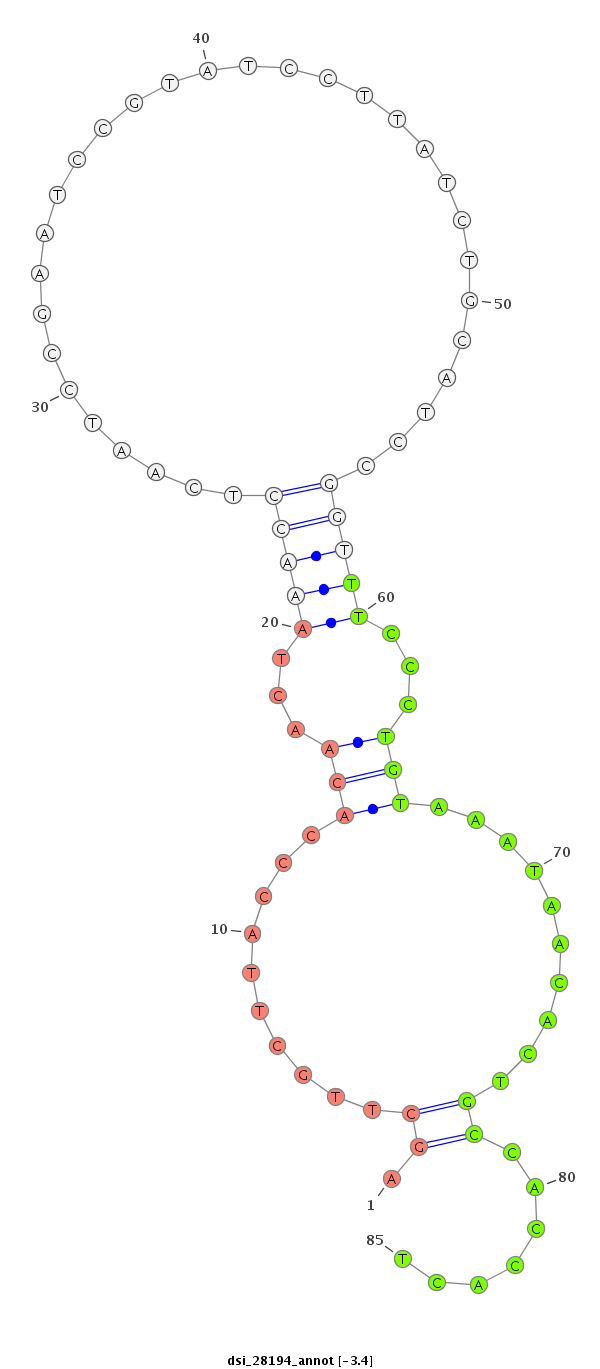
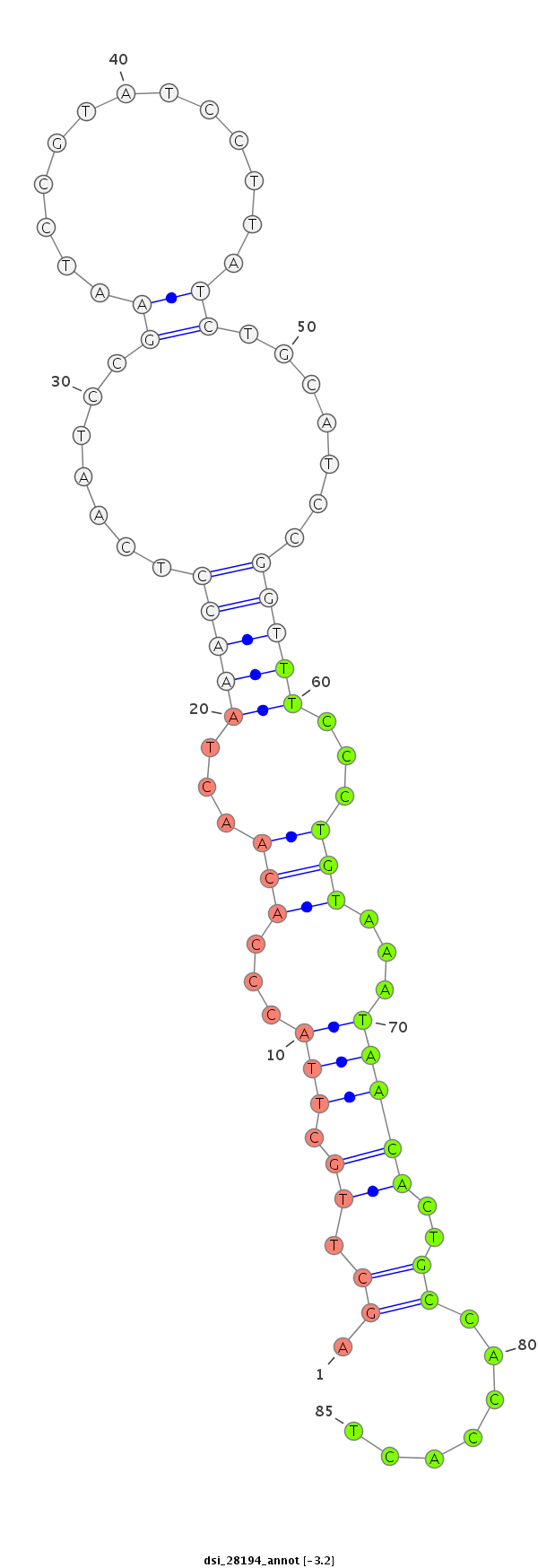
dsi_28194 |
CACAGCCACCA---GTTC---CTCCTCATC------------GGCATCGGGTCAATCCGGTGAAGAAGGTAAGCCAGAGCGGC--------------------------------------------------------TCC------------------CG---TCAG-C---------------------C-----------------AG-------CAGCCGCCCC---TCAGCTTGCT---TACCCACA------------------------------AC--------TAAACCTCAATCCG------A----------ATCCG--------------------TAT------CCTTATCTGCATCCGGTTT---------CCCTGTAAATAACA---------------CTGCCACCAC------------------TGAAACG---------------------------------TCTGC------CAAGAGCACCCTTAATCACCA-----------CATA------------G-----------------CCACCAAG------------TCAGCGGGC---AGCATATCAAGTGTAAGGGAG------ |
| droSec2 |
scaffold_77:93752-94001 - |
|
CACAGCCACCA---GTTC---CTCCTCATC------------GGCATCGGGTCAATCCGGTGAAGAAGGTAAGCCAGAGCGGC--------------------------------------------------------TCC------------------CG---TCAG-C---------------------C-----------------AG-------CAGCCGCCCC---TCAGCTTGCT---TACCCACA------------------------------AC--------TAAACCTCAATCCG------A----------ATCCG--------------------TAT------CCTTATCTGCATCCGGTTT---------CCCTGTAAATAACA---------------CTGCCACCAC------------------TGAAACG---------------------------------TCTGC------CAAGAGCACCCTTAATCACCA-----------CATA------------G-----------------CCACCAAG------------TCAGCGGGC---AGCATATCAAGTGTAAGGGAG------ |
| dm3 |
chr3R:14844156-14844405 + |
|
CACAGCCACCA---GTTC---CTCCTCATC------------GGCATCGGGTCAATCCGGTGAAGAAGGTAAGCCAGAGCGGC--------------------------------------------------------TCC------------------CG---TCAG-C---------------------C-----------------AG-------CAGCCGCCCC---TCAGCTTGCT---TACCCACA------------------------------AC--------TAAATCTCAATCCG------A----------ATCCC--------------------TAT------CCTTATCTGCATCCGGTTT---------CCCTGTAAATAACA---------------CTGCCACCAC------------------TGAAACG---------------------------------TCTGC------CAAGAGCACCCTTAATCACCA-----------CATA------------G-----------------CCACCAAG------------TCAGCGGGC---AGCATATCAAGTGTAAGGGAG------ |
| droEre2 |
scaffold_4770:6785169-6785424 - |
|
CACAGCCACCA---GTTC---CTCCTCATC------------GGCATCGGGTCACTCCGGTGAAGAAGGTAAGCCAGAGCGGC--------------------------------------------------------TCC------------------CG---TCAG-C---------------------C-----------------AG-------CAGCCGCCCC---TCCTCTTGCT---TACCCACA------------------------------AC--------TAAATCTCAATCCGAATCCGA----------ATCCG--------------------TAT------CCTTATCTGCATCCGGTTT---------CCTTGTAAATAACA---------------CTGCCACCAC------------------TGAAACG---------------------------------TCTGC------CAAGAGCACCCTTAATCACCA-----------CATA------------G-----------------CCACCAAG------------TCAGCGGGC---AGCATATCAAGTGTAAGGGAG------ |
| droYak3 |
3R:3522191-3522440 + |
|
CACAGTCACCA---GTTC---CTCCTCATC------------GGCATCGGGTCACTCCGGTGAAGAAGGTAAGCCTGAGCGGC--------------------------------------------------------TCC------------------CG---TCAG-C---------------------C-----------------AG-------CAGCCGCCCC---ACTGCTTTCC---TACCCACA------------------------------AC--------TAAACATCAGTCCG------A----------ATCCG--------------------TAT------CCTTATCTGCATCCAGTTT---------TCTTGTAAATAACA---------------CTGCCACCAC------------------TGAAACG---------------------------------TCTGT------CAAGAGCACCCTTAATCACCA-----------CATA------------G-----------------CCACCAAG------------TCAGCGGGC---AGCATATCAAGTGTAAGGGAG------ |
| droEug1 |
scf7180000409804:1084389-1084629 + |
|
CACAGCCACCA---GTTC---CTCCTCGTC------------AGCATCGGGCCACTCCGGTGAAGAAGGTAAGCCAGAGCAGC--------------------------------------------------------TCC------------------AG---TCAG-T---------------------C-----------------AG-------CAGCTGCCCC---ACCGCTTGCCAACCACCCACA------------------------------GT--------CAAACCACAA------------------------------------------------T------CCCTATCCGCATCCTGTTT---------CCCTGTAAATAACA---------------CCGCTACCAC------------------TGAAACG---------------------------------TCTGT------TAAGAGCACCCTTAACCACCA-----------TATA------------G-----------------CCAGCAAG------------TCAGTGGGC---AGCATATCAAGTGTGAGGGAG------ |
| droBia1 |
scf7180000302402:596709-596943 + |
|
CACGGCCACCA---GTTC---CTCCTCATC------------GGCATCGGCTCACTCCGGTGAAGAAGGTAAGCCAGAGCAGC--------------------------------------------------------TCC------------------AG---TCAG-C---------------------C-----------------AG-------CAACTGCTCGAAAACCGCTTGCC---AACC--------------------------------------------------TGAACCCG------CATCCATATCC-C---TGTCCG--------------CAA------CCCTATCCACATCCTGTAT---------CCCTGTAAATAACA---------------CTG---------------------------------------------------------------------A------CAAGAGCACCCTTAATCACCA-----------CACA------------G-----------------CCAGCAAG------------TCAGTGGGC---AGCATATCAAGTGTGAGGGAG------ |
| droTak1 |
scf7180000415256:338919-339138 + |
|
CACCGCCACCA---GTTC---CTCCTCATC------------GGCATCGGGTCACTCCGGTGAAGAAGGTAAGCCAGAGCAGC--------------------------------------------------------TCC------------------CG---TCAG-G---------------------C----GTC----------AG-------CA-------CACCTC------------------------------------------------------------------CAA-----------------------TCG--------------------TTT------CCCTATCCGCATCCTGTAT---------CCCTGTAAATAACA---------------CCGCCACCAC------------------TGAAACG---------------------------------TCTGC------CAAGAGCACCCTTAATCAGCA-CAG-------CACA------------G-----------------CCAGCAAG------------TCAGTGGGT---AGCATATCAAGTGTGAGGGAT------ |
| droEle1 |
scf7180000490994:598180-598414 + |
|
CACGGCCACCA---GTTC---CTCCTCATC------------GGCATCGGGCCACTCCGGTGAAGAAGGTAAGCCAGAGCAGC--------------------------------------------------------TCC---------------GCCCG---TCAG-C---------------------C-----------------AG-------CAGCTGCCCC---ACC---TCCG---TACCCGTA--------------------------------------------------TCCA------T----------ATCTGTATCCTTATCCAC--AGCCACAT------CCACATCCACATCCTGTTC---------CCTTGTAAATAACA---------------CTGCTATGA---------------------------------------------------------------G------CAAGAGCACCCTTAACCAGCATCAGCA-----CATA------------G-----------------CCAGCAAG------------------------------TCAAGTGTGGGGGAG------ |
| droRho1 |
scf7180000779900:495742-495994 + |
|
CACGGCCACCA---GTTC---CTCCTCATC------------GGCATCGGGCCACTCCGGTGAAGAAGGTAAGCCAGAGCAGC--------------------------------------------------------ATC------------------CG---TCAG-C---------------------C-----------------AG-------CAGCTGCCCCACCTCCGCTTGCC---CACCCACA------------------------------AT--------C------CAGCCCG------T----------ATCCC--------------------TAT------CCACATCCACATTCTGTTC---------CCTTGTAAATAACA---------------ATGCTAGCAC------------------AGCAACG---------------------------------TCTGCTACGAGCACGAGCACCCTTAATCAGCA-----------CATA------------G-----------------CCAGCAAG------------TCAGCGGGC---AGCATATCGAGTGTGAGGAGG------ |
| droFic1 |
scf7180000453812:196118-196325 - |
|
GACGGCCACCA---GTTC---CTCCTCGTC------------GGCATCGGGCCACTCCGGTGAAGAAGGTAAGCCAGAGCAGC--------------------------------------------------------CAG------------------CG---ACGG-A---------------------G-----------------CA-------GAGCTGCCCC---ACCGCTCGCC---CACCCAAC------------------------------AC--------------------------------------------------------------------------------CCCGTCCTGTCC---------CCCTGTAAATAACA---------------CCGA---------------------------------------------------------------------------AAACAGCACCCTTAATCAGCA-----------CATA------------G-----------------CCAGCAAG------------TCAGTGGGC---AGCATATCAAGTGTGAGGGAGCAGCAT |
| droKik1 |
scf7180000302778:97651-97933 - |
|
CACGGCCACCA---GTTC---CTCCTCATC------------GGCCTCGGGCCACTCCGGCGAAGAAGGTAAGCCCGAGCAGC--------------------------------------------------------AAC------------------CG---TCAG-C---------------------C----GTC----------AG-------CAGCTGCCCC---TTCGCTTGCC---CAACCAAG------------------------------AC--------CCAACCCGAACCCA------A----------AACCA--------------------CAT------CCTCAATCACGTCCCGCTC---------CCTTGTAAATAACACGCTACTTTGCAACACTGTCAGCAC------------------AGCAACTCTAACAGCAGCAGCC---------------G---CATC------CAAGAGCACCCCCAATAACCA-----------TGTA------------G-----------------TCAGCAAG------------TCAGGGGGC---AGCATATCAAGTGTAAGGGAG------ |
| droAna3 |
scaffold_12911:3204573-3204801 + |
|
CGCAGCCACCAATAGTTC---CTCC------------------------------TCCGGCGAAGAAGGTAAGCCCGCGCAGC--------------------------------------------------------ACC------------------AGCT-TCAT-C---------------------C-----------------AGACAAAATGGGCTGCCGA---AGC-----------------------------------------------------------------------G------T------ATCC-C---AAACCG--------------TAACCCAAGTCCAAACCACGTCCTGTTC---CTTTGTCCCTGTAAATAACACGCTACTTTGCAACAC------------------------------------------------------------------------------TAAGAGCACCCTG-----CCA-----------CATA-ACCATCCAGAAA-----------------GCAGCAAG------------TCAGCGGGC---AGCATATCAAGTGTGAGG--------- |
| droBip1 |
scf7180000396640:78425-78662 + |
|
CGCAGCCACCAACAGTTC---CTCC------------------------------TCCGGCGAAGAAGGTAAGCCCGCGCAGC--------------------------------------------------------ACC------------------AGCT-TCAT-C---------------------C-----------------AGAC-----CAACCG-------ATGGCCTGCC-----------------------------------------------------------AAACCG------A------GTCC-CCCGTAACCG--------------TAACCCAAGTCCCAACCAAGCCCTGTTC---CTTTGTCCCTGTAAATAACACGCTACTTTGCAACAC------------------------------------------------------------------------------TAAGAGCACCCTG-----CCA-----------CACA-ACCGTTCAGAAG-----------------GCAGCAAG------------CCAGCGGGC---AGCATATCAAGTGTGAGGCAG------ |
| dp5 |
2:14967464-14967791 + |
dps_563 |
CACGGCCACCA---ATTCGTCCTCCTCTAC------------GGCGTCGGCCCACTCCGGCGAAGAAGGTAAGCTAGTGCTCC--------------------------------------------------------AGC------------------CG---CCGG-C---------------------A-----------------GA-------CAGTTGCCAT---TTCGCTTGCC---CACCCACACATACACCTACACCTACACCTACACTTAAGTC--------TAATCCCCAAACCG------A----------AT--------------------------------------CCAAACCCTGTTCCTCCTCTGCTAATGTATATAACACGCTACTTTGCAACACTGCCACAAC------------------AGAAGCA---GCAGCAACAGCAACAG------------------------CAACAAA---CA----------TTAACAAACAC----TACAATTTTGGAGTCAA--------------AAGCAAGTCAAAATCCGCATCAATGGGCACGAGCATATCAAGCGTAGGG--------- |
| droPer2 |
scaffold_0:5442550-5442874 - |
|
CACGGCCACCA---ATTCGTCCTCCTCTAC------------GGCGTCGGCCCACTCCGGCGAAGAAGGTAAGCTAGTGCTCC--------------------------------------------------------AGC------------------CG---CCGG-C---------------------A-----------------GA-------CAGTTGCCAT---TTCGCTTGCC---CACCCACACATACACCTA------CACCTACACTTAAGTC--------TAATCCCCAAACCG------A----------AT--------------------------------------CCAAACCCTGTTCCTCCTCTGCTAATGTATATAACACGCTACCTTGCAACACTGCCACAAC------------------AGAAGCA---GCAGCAACAGCAACAG------------------------AAACAAA---CA----------TTAACAAACAC----TACAATTTCGGAGTCAA--------------AAGCAAGTCAAAATCCGCATCAATGGGCACGAGCATATCAAGCGTAGGGCAG------ |
| droWil2 |
scf2_1100000004902:8232743-8233103 - |
dwi_3145 |
GACGGCCACCA---ATTC---ATCATCAT------------------CTGGAAATTCTGGGGAAGAAGGTAAGCAGCAGCAGC---------------AAGATGAAAATGAAGAA--------------------------------GAGGAGGAGGAGG---AGAAAGAGAA------------------GATGCAAAAAGATAATTTT----AATGCCAATTGCAAT---TTTGCTTGTTCCTTGCCCAAA------------------------------TCCTCCAAATC---TCCCAA---------------------------------------------------------TTG--TCCCCCTTGTAT---CCCTGCTAATGTAAATAACACGCTACTTTGCAACACTGCCACTACACACAATACAACAACAAAAACAACA---------------ACAGAAGCAACAACATCATCTAC------TATGTGTACTATTAACAACAA-----------C---AACAATA--AGA----A--------------TAGCAACACAAAATTCGCATCAATGAGC---AGCATATCAAGTGTAGAGAAG------ |
| droVir3 |
scaffold_13047:12984183-12984552 - |
dvi_16713 |
CACGGCAACCA---GTTC---ATCATCGGC---CACAACTACCGCCTCGGCTAGCTCTGCGGAAGAAGGTAAGAATCTGCGAACAGCAACCCAACAACAACAACAAGAACAACAACA--------GCAACTGCAACAACAACAACCAGAGCAACAACAACTG---CCACAT---------------------CAGCAGCAACAACAATTG----AATGCCAGTTGCAGT-----------------------------------------------------------------------------G------A------T----TTTGTTAACGTAACCGCTTTGACGCA-------CCCAAACCAAATCCT----------TGCTATTGTAAATAATACGCTACTACACTCTACTACCACAAC------------------AACAACA---------------------------------ACTTT------TAACAACAACAACAACAACAA--------------------CA--ACAGCCACTCAAATTTGTTGCTGAGCAAGTCAAAATTCGCATCAATGAGC---AGCATTTCATGTGTA------------ |
| droMoj3 |
scaffold_6540:28869645-28870019 + |
dmo_2106 |
CACGGCCACCA---ACTC---ATCATCATCGACGGCGACGACGGCGTCAGTCAGTTCCGTGGAAGAAGGTAAGGCAACA-----------------ACGACAACAACAACAACAACGACACAACTACAACTACAACAACTACTGTCAGAGCAACAGCAGC---TGCAACAGCAACGACAACAACAACAACAACAGCAACAACAACCATTA----AATTCTAGCTGCAAT---AA-----------------------------------------------------------------------CT------T----------ATCCG--------------------CTT------TGTTAGACCCAAACCTTGC---------CATTGTAAATAATACGCTACTACACTCTACTACCACATC------------------AACAACA---ACAGCAACAACA---------------CCAACTTT------TAACGTCATGAACAACAACAA-----------C---AACAACA--AGAGTCAAACAAATTTGTTGCTCAGCAAGTCAAAATTC-------------------------GCATCAATGAG------ |
| droGri2 |
scaffold_15074:4005000-4005064 - |
|
GACGGCAACCA---ACTCATCCAGTTCGAC------------AGCCTCTGCACACTCTACGGAAGAAGGTAAGATGGAAC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |