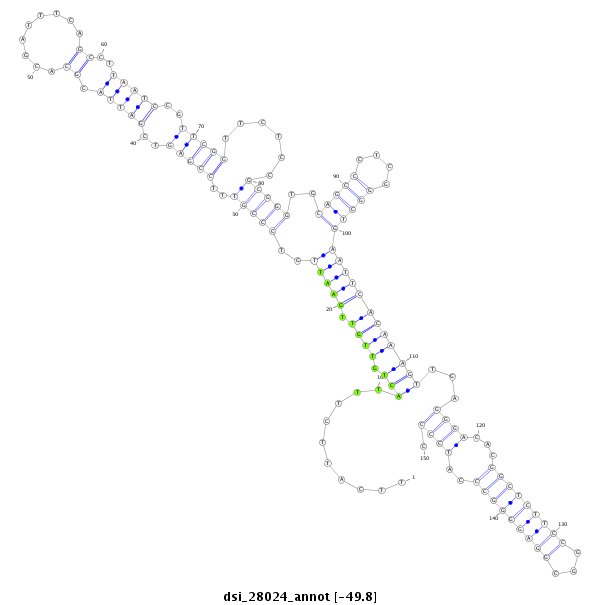


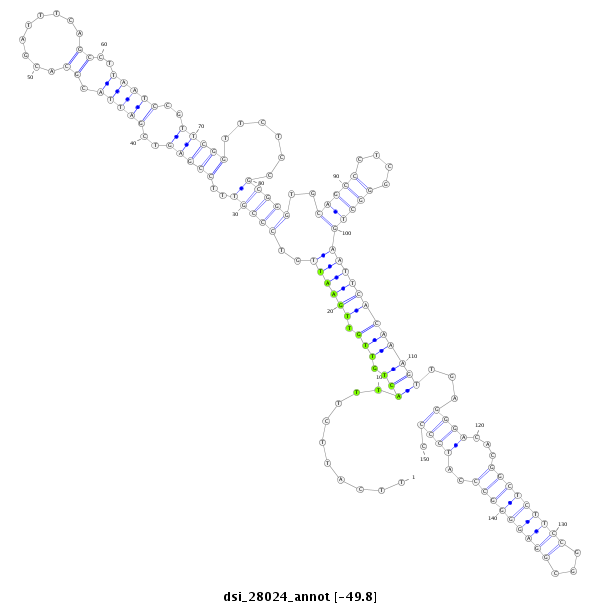
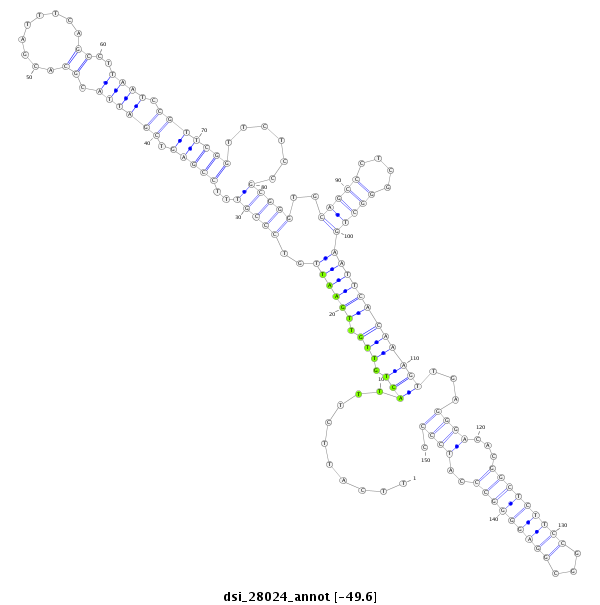
ID:dsi_28024 |
Coordinate:3l:22625380-22625529 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -49.8 | -49.6 | -49.6 |
 |
 |
 |
intergenic
No Repeatable elements found
| mature | star |
|
ATGTCTGCCTGCGTCACCTATGTTCCGCAGCGCGTACTTTAGCGAGTTTTTTCATTCTTTACTGTTGTTGAATTGTCCCGTTTCCGAGTCGATTACGCACGATTTCAGCCTTAATCCGTTCGGTTCTCCGCGGGTGCAGCCCTCGGGCTGAATTCACAAAGTTGAGGGACACGGCTCTTCCGGCGGAGGGGCCCATCCCCCTGCCGATTGGGCTTCCCGGGACAGTCGCCGAAGCGTAACCGAAAACTAT
**************************************************..........(((.((((.(((((..(((((..(((((..(((((.((.........))..)))))..)))))......)))))..(((((....)))))))))))))))))...((((...(((((((((...)))))))))..)))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
M024 male body |
SRR553485 Chicharo_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................GATTGGTCTTCCAGGGAC........................... | 18 | 2 | 4 | 1.00 | 4 | 3 | 1 | 0 | 0 |
| ..........................................................TTACTGTTGTTGAAT................................................................................................................................................................................. | 15 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................................................................ACGATTGGTCTTCCAGGGAC........................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................CCGCGGGTGCAGATCTTGG........................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
|
TACAGACGGACGCAGTGGATACAAGGCGTCGCGCATGAAATCGCTCAAAAAAGTAAGAAATGACAACAACTTAACAGGGCAAAGGCTCAGCTAATGCGTGCTAAAGTCGGAATTAGGCAAGCCAAGAGGCGCCCACGTCGGGAGCCCGACTTAAGTGTTTCAACTCCCTGTGCCGAGAAGGCCGCCTCCCCGGGTAGGGGGACGGCTAACCCGAAGGGCCCTGTCAGCGGCTTCGCATTGGCTTTTGATA
**************************************************..........(((.((((.(((((..(((((..(((((..(((((.((.........))..)))))..)))))......)))))..(((((....)))))))))))))))))...((((...(((((((((...)))))))))..)))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR902009 testis |
SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
GSM343915 embryo |
SRR553488 RT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................TAACTGGGCAAAGGCT................................................................................................................................................................... | 16 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................AAGGGCCAAGTCAGCGG.................... | 17 | 2 | 12 | 0.33 | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................AGGGGTACAGCTAACCC...................................... | 17 | 2 | 9 | 0.22 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ........................................................................................................AGTCCAAATTAGGCAAGC................................................................................................................................ | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................................................................................GGCTAACTCGATGGGGCCT............................ | 19 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................GGAGATGGCTAACCCGA.................................... | 17 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................CACGTCGGGAGCCCA...................................................................................................... | 15 | 1 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................GGCAAGCCAGGAGGTGC...................................................................................................................... | 17 | 2 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................AATTAGCCAAGCCTGGAGG......................................................................................................................... | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................AAGTCGCAAATAGGCAAGGC............................................................................................................................... | 20 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................ACGGCTAACCAGAAAG................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................AGCCCAGTGGCTCCCACG................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................GGCGAAACCAAAGGGCCCT............................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................GAAGGCCGCTTGCCCG.......................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 3l:22625330-22625579 + | dsi_28024 | ATGTCTGCCTGCGTCACCTATGTTCCGCAGCGCGTACTTTAGCGAGTTTTTTCATTCTTTACTGTTGTTGAATTGTCCCGTTTCCGAGTCGATTACGCACGATTTCAGCCTTAATCCGTTCGGTTCTCCGCGGGTGCAGCCCTCGGGCTGAATTCACAAAGTTGAGGGACACGGCTCTTCCGGCGGAGGGGCCCATCCCCCTGCCGATTGGGCTTCCCGGGACAGTCGCCGAAGCGTAACCGAAAACTAT |
| droSec2 | scaffold_1133:5656-5715 - | --------------TA-------------------------TCGGGTCTCTTCATTCTTAACGACACTTTATTTTTTTTATTTTCGATTCGTTTTCGCA------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEre2 | scaffold_4842:78368-78405 + | --------------CACCTGTGTTACGCAGCGCAGATTTTAGCAAACCTTTT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droEle1 | scf7180000486278:4765-4774 + | GTGTCTGACT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
Generated: 05/18/2015 at 07:33 PM