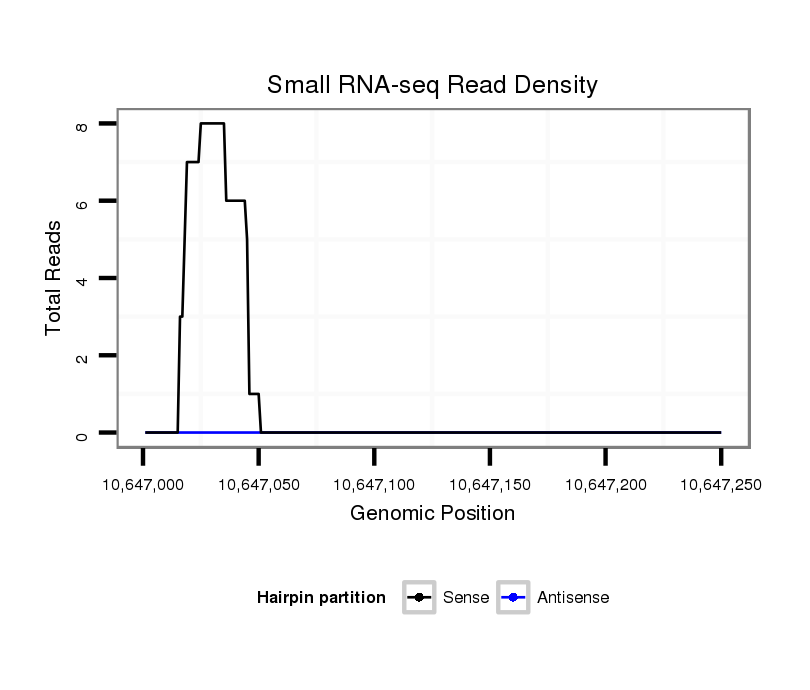
ID:dsi_2774 |
Coordinate:3r:10647051-10647200 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
intergenic
No Repeatable elements found
|
GGGCAGAGATCCACCTGCACAATGTTCAGCCGGTCCAGTTGGAGATGATTGTAAGTATTGCTAATGTTATTCAACATCTTGATGAGTCAGTCGTATTATGTTGGGGGGCTTCAACCCAAGTTTTGATTTCACCATATTGTGTGGGAGACAAGACTTGGTTATGTGAAATATTTCAGAGTTGCCAAGTCTGAGTCAGTAGGAGCATATATTGTATATTTATATATGTATGCTAGGAATAGGAACTATATAG
**************************************************.....(((((.....(((.((((((((....((((.....))))...)))))))).)))((((..((((((((((.((((.(((((...))))))))))))))))))).....))))....((((((.((...)).)))))).)))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
M025 embryo |
SRR553485 Chicharo_3 day-old ovaries |
M053 female body |
SRR553488 RT_0-2 hours eggs |
M024 male body |
GSM343915 embryo |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............TGCACAATGTTCAGCCGGTCCAGTTGGAGA............................................................................................................................................................................................................. | 30 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................CACAATGTTCAGCCGGTC....................................................................................................................................................................................................................... | 18 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................ACAATGTTCAGCCGGTCCAGTTGGAG.............................................................................................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................ACAATGTTCAGCCGGTCCAGTTGGAGA............................................................................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................TTCAGCCGGTCCAGTTGGAGATGATT........................................................................................................................................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................GTTCAGCCGGTCCAGTTGGAGAC............................................................................................................................................................................................................ | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...CTAGGATCCACCTGCACAATGTTC............................................................................................................................................................................................................................... | 24 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................GGAGAGCATTGTAATTATTGCT............................................................................................................................................................................................ | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................TTTGAGTCAATAGGAGGAT............................................. | 19 | 3 | 16 | 0.31 | 5 | 1 | 0 | 0 | 0 | 4 | 0 | 0 | 0 |
| .......................................................................................................................................ATTGTGTGGGAGGCAGGACTC.............................................................................................. | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................GTTATTCAAAGTCGTGATGA..................................................................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................AGAGACAAGGCTTGGTTGT........................................................................................ | 19 | 3 | 14 | 0.14 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| .........................................................................................................................................................................................................................TACATAGGTATGCTAGG................ | 17 | 2 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
|
CCCGTCTCTAGGTGGACGTGTTACAAGTCGGCCAGGTCAACCTCTACTAACATTCATAACGATTACAATAAGTTGTAGAACTACTCAGTCAGCATAATACAACCCCCCGAAGTTGGGTTCAAAACTAAAGTGGTATAACACACCCTCTGTTCTGAACCAATACACTTTATAAAGTCTCAACGGTTCAGACTCAGTCATCCTCGTATATAACATATAAATATATACATACGATCCTTATCCTTGATATATC
**************************************************.....(((((.....(((.((((((((....((((.....))))...)))))))).)))((((..((((((((((.((((.(((((...))))))))))))))))))).....))))....((((((.((...)).)))))).)))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
M024 male body |
|---|---|---|---|---|---|---|---|
| ........................................................................................................................................ACCACGGCCTCTGTTCTGAAC............................................................................................. | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 |
| ........................................................................................................................................................................................TCAGACTCGGTCGTCCTCG............................................... | 19 | 2 | 4 | 0.50 | 2 | 0 | 2 |
| ....................TTACGAGTCGGGCAGATC.................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 |
Generated: 04/24/2015 at 10:46 AM