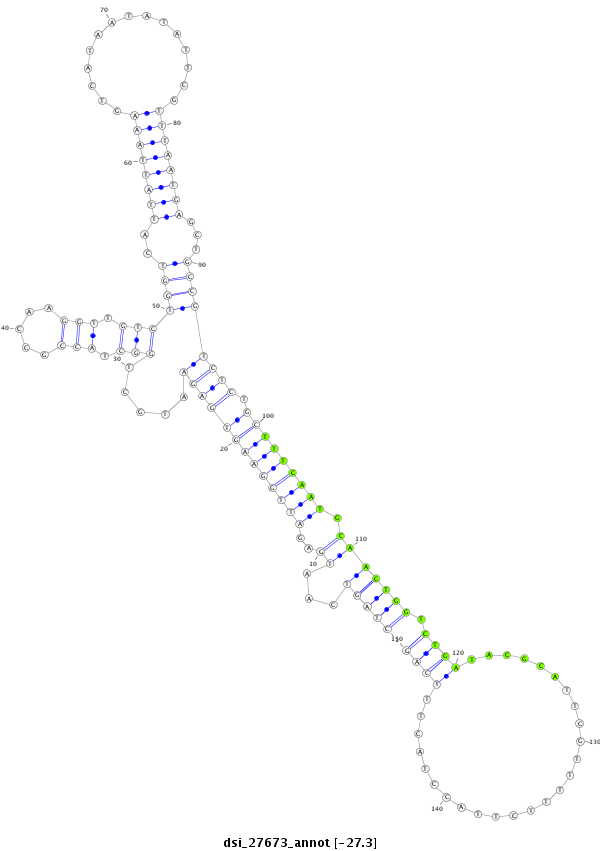

ID:dsi_27673 |
Coordinate:3r:19919182-19919331 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -27.2 | -27.0 | -27.0 |
 |
 |
 |
five_prime_UTR [3r_19919082_19919181_-]; exon [3r_19917129_19919181_-]; intron [3r_19919182_19932503_-]
No Repeatable elements found
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CACCATCGGAATTGGGCTGAACTTAAAATGCCCCATTGGAAAACCAGTAACTAGTCAATGAGATTGGAAGTGAGAATGCTGGCTACCGCCAAGGTTGTCTGGTCATTATTAAAGTCATAATATATTCGTTTAATGAGCTGCCGTCTCTGCTTTCAATGCAACTGGTCTGATACGCATTCGTTTTTCTTACCTACTTTCAGATACTACAAGATAAAGTCGGGCCTGGGCGCCCATCTAGAGAGGGAGAGGA **************************************************(((((...((..(((((((((((((.....(((.(((.....))).)))((((..((((((((...............))))))))...)))))))).))))))))).))))))).((((..........................))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
M053 female body |
SRR553488 RT_0-2 hours eggs |
M023 head |
SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
M025 embryo |
SRR902008 ovaries |
SRR618934 dsim w501 ovaries |
SRR902009 testis |
SRR1275487 Male larvae |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................TTAGAAGTGAGAATGCCG......................................................................................................................................................................... | 18 | 2 | 3 | 11.67 | 35 | 20 | 4 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................TTAGAAGTGAGAATGCCGG........................................................................................................................................................................ | 19 | 2 | 1 | 7.00 | 7 | 5 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................TTAGAAGTGAGAATGCCGGT....................................................................................................................................................................... | 20 | 3 | 4 | 6.50 | 26 | 13 | 7 | 0 | 4 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................................................................TAGAAGTGAGAATGCCGG........................................................................................................................................................................ | 18 | 2 | 1 | 5.00 | 5 | 3 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................GCTTAGAAGTGAGAATGCCGG........................................................................................................................................................................ | 21 | 3 | 1 | 4.00 | 4 | 1 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................GCTTAGAAGTGAGAATGCCG......................................................................................................................................................................... | 20 | 3 | 6 | 2.50 | 15 | 10 | 2 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................GAAGTGAGAATGCCGG........................................................................................................................................................................ | 16 | 1 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................AAGTGAGAATGCTGAC....................................................................................................................................................................... | 16 | 1 | 2 | 1.50 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................ATCGGAAGTGAGAATGC........................................................................................................................................................................... | 17 | 1 | 2 | 1.50 | 3 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................CTTAGAAGTGAGAATGCCG......................................................................................................................................................................... | 19 | 3 | 17 | 1.35 | 23 | 15 | 1 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................ACAAGATAAAGTCGGGCC........................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................GATAAGATACTACAAGATAAAGTCGGG............................. | 27 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TAGAAGTGAGAATGCTGGT....................................................................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................TCGGAAGTGAGAATGCTG......................................................................................................................................................................... | 18 | 1 | 2 | 1.00 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................ACAAGATAAAGTCGGGCCTGGGCGC.................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................ACTACAAGATAAAGTCGGGCCT.......................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................TTTCAATGCAACTGGTCTGATACGCA.......................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................ATCGGAAGTGAGAATGCTG......................................................................................................................................................................... | 19 | 1 | 2 | 1.00 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................AGTGAGAATGCTGGCA...................................................................................................................................................................... | 16 | 1 | 2 | 1.00 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................TTGGAAGTGCGAATGCTG......................................................................................................................................................................... | 18 | 1 | 2 | 1.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................TCGGAAGTGAGAATGCT.......................................................................................................................................................................... | 17 | 1 | 2 | 1.00 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................TTAGAAGTGAGAATGCCGGG....................................................................................................................................................................... | 20 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................TTGGAAGTGAGAATTCT.......................................................................................................................................................................... | 17 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................ATCGGAAGTGAGAATG............................................................................................................................................................................ | 16 | 1 | 4 | 0.75 | 3 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TAGAAGTGAGAATGCCGGT....................................................................................................................................................................... | 19 | 3 | 12 | 0.75 | 9 | 4 | 4 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................GCTTAGAAGTGAGAATGC........................................................................................................................................................................... | 18 | 2 | 4 | 0.75 | 3 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................GAAGTGAGAATGCCGGT....................................................................................................................................................................... | 17 | 2 | 7 | 0.57 | 4 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................AAGTGAGAATGCCGG........................................................................................................................................................................ | 15 | 1 | 7 | 0.57 | 4 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................CTTAGAAGTGAGAATGCCGG........................................................................................................................................................................ | 20 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................GAAGTGAGAATGCTGAC....................................................................................................................................................................... | 17 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................ATCGGAAGTGAGAATGCT.......................................................................................................................................................................... | 18 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TGGAAGTGTGAATGCT.......................................................................................................................................................................... | 16 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................TTAGAAGTGAGAATGC........................................................................................................................................................................... | 16 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................TCGGAAGTGAGAATGC........................................................................................................................................................................... | 16 | 1 | 8 | 0.38 | 3 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................CGGAAGTGAGAATGCTG......................................................................................................................................................................... | 17 | 1 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................GTGCTTAGAAGTGAGAATGC........................................................................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................GGAAGTGAGAATGCTGA........................................................................................................................................................................ | 17 | 1 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................GGAATTGAGAATGCTGAC....................................................................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................TGGGTACCGCGAAGGTTGCC....................................................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................AGTGAGAATGCTGAC....................................................................................................................................................................... | 15 | 1 | 12 | 0.25 | 3 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................CTTAGAAGTGAGAATGCT.......................................................................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................CGGAAGTGAGAATGC........................................................................................................................................................................... | 15 | 1 | 14 | 0.21 | 3 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TGGAAGTGCGAATGCTG......................................................................................................................................................................... | 17 | 1 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................TTGGAAGTGCGAATGC........................................................................................................................................................................... | 16 | 1 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................GAAGTGAGAATGCCGGTA...................................................................................................................................................................... | 18 | 3 | 20 | 0.20 | 4 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................ACTGGAAGTCAGAATGCT.......................................................................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................ATCGGAAGTGAGAATTCT.......................................................................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................GCAAGTGAGAATGCTG......................................................................................................................................................................... | 16 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................AAGTGAGAATGCTGA........................................................................................................................................................................ | 15 | 1 | 13 | 0.15 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................CGCTTAGAAGTGAGAATGC........................................................................................................................................................................... | 19 | 3 | 14 | 0.14 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................TCGGAAGTGAGAATG............................................................................................................................................................................ | 15 | 1 | 14 | 0.14 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................CGCTTAGAAGTGAGAATGCT.......................................................................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................AGTGAGAATGTTGGC....................................................................................................................................................................... | 15 | 1 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................ATCTGAAGTGAGAATGCT.......................................................................................................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TAGAAGTGAGAATGCCGGG....................................................................................................................................................................... | 19 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................AGAAGTGCGAATGCTGGC....................................................................................................................................................................... | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................TCGGAAGTGAGAATTCTG......................................................................................................................................................................... | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................AAGTGAGAATGTCGGCT...................................................................................................................................................................... | 17 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................GGAAGTTAGAATGCTGA........................................................................................................................................................................ | 17 | 2 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................ATCGGAAGTGAGAAT............................................................................................................................................................................. | 15 | 1 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................ATCGGAAGTGAAAATGCT.......................................................................................................................................................................... | 18 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................GGAAGCGAGAATGTCGGCT...................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................GCGCTTAGAAGTGAGAAT............................................................................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
GTGGTAGCCTTAACCCGACTTGAATTTTACGGGGTAACCTTTTGGTCATTGATCAGTTACTCTAACCTTCACTCTTACGACCGATGGCGGTTCCAACAGACCAGTAATAATTTCAGTATTATATAAGCAAATTACTCGACGGCAGAGACGAAAGTTACGTTGACCAGACTATGCGTAAGCAAAAAGAATGGATGAAAGTCTATGATGTTCTATTTCAGCCCGGACCCGCGGGTAGATCTCTCCCTCTCCT
**************************************************(((((...((..(((((((((((((.....(((.(((.....))).)))((((..((((((((...............))))))))...)))))))).))))))))).))))))).((((..........................))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
M023 head |
M053 female body |
M025 embryo |
SRR553485 Chicharo_3 day-old ovaries |
GSM343915 embryo |
SRR618934 dsim w501 ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR1275487 Male larvae |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................CGGGGCAACCTTTAGGTCA.......................................................................................................................................................................................................... | 19 | 2 | 2 | 48.00 | 96 | 60 | 26 | 7 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................CGGGGCAACCTTTAGGTCAT......................................................................................................................................................................................................... | 20 | 2 | 1 | 16.00 | 16 | 7 | 6 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................CGGGGCAACCTTTAGGTCATG........................................................................................................................................................................................................ | 21 | 3 | 4 | 16.00 | 64 | 41 | 10 | 11 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................CGGGGCAACCTTTAGGTC........................................................................................................................................................................................................... | 18 | 2 | 5 | 8.80 | 44 | 26 | 9 | 5 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 |
| ...........................TCCGGGGCAACCTTTAGGTCA.......................................................................................................................................................................................................... | 21 | 3 | 3 | 3.67 | 11 | 0 | 0 | 8 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 |
| ..........................TTCCGGGGCAACCTTTAGGTCA.......................................................................................................................................................................................................... | 22 | 3 | 2 | 3.00 | 6 | 1 | 0 | 4 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................TTTCCGGGGCAACCTTTAGGTCA.......................................................................................................................................................................................................... | 23 | 3 | 1 | 3.00 | 3 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................CGGGGTAACCTTTAGGTCAT......................................................................................................................................................................................................... | 20 | 1 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................TTTCCGGGGCAACCTTTAGGTCAT......................................................................................................................................................................................................... | 24 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................CGGGGTAACCTTTAGGTC........................................................................................................................................................................................................... | 18 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................TCCGGGGCAACCTTTAGGTCAT......................................................................................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................TTTCCGGGGCAACCTTTAGGTC........................................................................................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................CGGGGTAACCTTTAGGTCATG........................................................................................................................................................................................................ | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................CAGGGCAACCTTTAGGTCAT......................................................................................................................................................................................................... | 20 | 3 | 2 | 1.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................GGGGCAACCTTTAGGTCA.......................................................................................................................................................................................................... | 18 | 2 | 8 | 0.88 | 7 | 1 | 0 | 5 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................TTTCCGGGGCAACCTTTAGGT............................................................................................................................................................................................................ | 21 | 3 | 3 | 0.67 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................GGGGCAACCTTTAGGTCAT......................................................................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................GGGCAACCTTTAGGTCAT......................................................................................................................................................................................................... | 18 | 2 | 4 | 0.50 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................CCGGGGTAACCTTTAGGTC........................................................................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................TTCCGGGGCAACCTTTAGGTC........................................................................................................................................................................................................... | 21 | 3 | 4 | 0.50 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ...................................................................................ATGGCGGTTTCGAAAGACCAGT................................................................................................................................................. | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................CCGGGGCAACCTTTAGGTCAT......................................................................................................................................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................CAGAGACTAAAGTTACATTGT....................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................CGGGCCAACCTTTAGGTCAT......................................................................................................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................CAGGGCAACCTTTAGGTCA.......................................................................................................................................................................................................... | 19 | 3 | 14 | 0.14 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................CGGGGCAACCTTTAGGTTAT......................................................................................................................................................................................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................AGACTGGATGAAAGT................................................... | 15 | 1 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................TTTCCGGGGCAACCTTT................................................................................................................................................................................................................ | 17 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................AAAGAATGGATGTAA..................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................GGGGCAACCTTTAGGTC........................................................................................................................................................................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
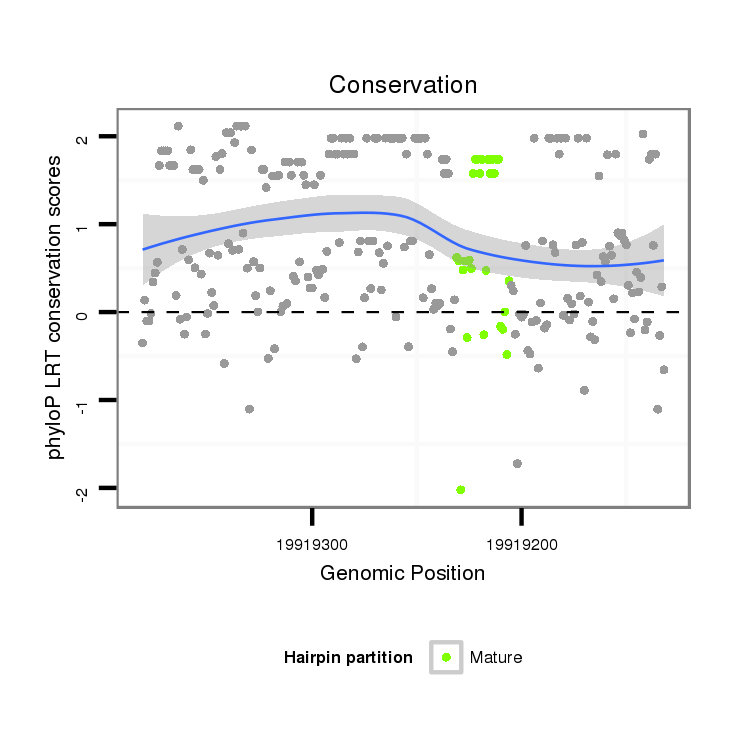
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 3r:19919132-19919381 - | dsi_27673 | CACCATCG--GAATTGGGCTGAACTTAA-AATGCCC----------------------------------------------CATTGGAAAACCAGTAACTAG--T-------------------CAATGAGATTGGAAG-TGAGAATGCTGGCTACC-----------------------GCCAAGGTTGTCTGGT----CATTA---TTAAAG-----TCATAATATATTCGTTTAATGAGCT--GCC---G--TCTCTGCTTT-CA------ATGCAACTG--G-----TCTGATACGCATTC-----GTTT-TTC---TTACCTACTTTCAGATACTA---C-A-A--GATA------------------AAGTCGGGCCTGG-------GCGCCCA--------------TCT----AGAGAGGGAG-------AGGA------- |
| droSec2 | scaffold_0:20723887-20724136 - | CACCATCG--GAATTGGGCTGAACTTAA-AATGCCC----------------------------------------------CATGGGAAAACCAGTAACTAG--T-------------------CAATGAGATTGGAAG-TGAGAATGCTGGCTACC-----------------------GCCAAGGTTGTCTGGT----CATTA---TTAAAG-----TCATAATATATTCGTTTAATGAGCT--GCC---G--TCTCTGCTTT-CA------ATGCAACTG--G-----TCTGATACGCATTC-----GTTT-TTC---TTACCTACTTTCAGATACTA---C-A-A--GATA------------------AAGTCGGGCCTGG-------GCGCCCA--------------TCT----AGAGAGGGAG-------AGGA------- | |
| dm3 | chr3R:20388615-20388866 - | CACCATCGGGGAATTGGGCTGAACTTAA-AATGACC----------------------------------------------CATTGGAAAACCAGTAACTAG--C-------------------CAATGAGATTGGAAG-TGAGAATGCTGGCTACC-----------------------ATCAAGGTTGTCTGGT----CAATA---TTAAAG-----TCATAATATATTCGTTTAATGAGCT--GCC---G--TCTCTGCTTT-CA------ATGCAACTG--G-----TCTGATTCGCATTC-----GTTT-TTC---TTACCTACTTTCAGATACTA---C-A-A--GATA------------------AAGTCGGGCCTGG-------GCGCCCA--------------TCT----AGAGAGGGAG-------AGGA------- | |
| droEre2 | scaffold_4820:7668592-7668841 + | CACCATCG--GAATTGGGCTGAACTTAA-AATGCTC----------------------------------------------CATTGG-AAACCAGTAACTAG--C-------------------CAATGAGACTGGAAG-TGAGAATGCTGGCTACC-----------------------GCCAAGGTTGTCTGGT----CATAA---TTAAAG-----TCATAATATATTCGTTTAATGAGCT--GAC---G--TCTCGATTTT-CA------ATGCAACTG--G-----TCTGATTCGCATTCTC---A-TT-TTT---CTACCTACTTTCAGATACTA---C-A-A--GATA------------------ACGTCGGGCCTGG-------GCGCCCA--------------TCT----AGAGAAGGAG-------GGGA------- | |
| droYak3 | 3R:26110598-26110855 + | CACCAACG--GAATTGGGCTGAACTTAA-AATGCTC----------------------------------------------CATTGGAAAACCAGTAACCAG--T-------------------CAATGAGATTGGAAG-TGAGAATGCTGGCTACC-----------------------GCCAAGGTTGTCTGGT----CATTA---TTAAAG-----TCATAATATATTCGTTTAATGAGCT--GAC---G--TCTCTGTTTT-CA------ATGCAACTG--G-----TCTGATTCGCATTCCC---A-TT-TGT---CTACCTACTTTCAGATACTA---C-A-A--TATA------------------ACGCCGGGCCTGG-------GCGCCCA--------------TCT----AGAGAAGGAGAGAGAGGAAGA------- | |
| droEug1 | scf7180000409881:111795-112051 - | TACTATCG--GAATTGGGCTAAACGTAG-AATGTCTCAATCG-----------------------------------CTGGCCATTGGGAAACCAGTAACCAG--T-------------------CAATGAGATTGGAAA-TGAGAATGCTGGCTGCC-----------------------GCCAAGGTTGTCTGGC----CATTA---TAAAAG-----TCATAATTTATTCATTTAATGAGAA--GCC---GTCTCTCTGTTTC-CA------CTGCAACTG--G-----TCTGATTTGA-------TTTGTCTTGT---TTTTCT-CTTTCAGATACCA---C-A-A--GACA------------------ACATCGGGCCTGG-------GCGCCCA--------------TCT----AGAGAAGGAG------------------ | |
| droBia1 | scf7180000302402:4163240-4163506 - | CACTATCG--GAATTGGGCTGAGCGTAA-AATGTCTCAATAG-----------------------------------ATGGCCATTGGCAAACCAGTCACCATAGTGACT----------ATAGTCAATGGGATTGGAAA-TGAGAATGCTGGCTGCC-----------------------GCCAAGGTTGTCTGGT----CTCTA---CAGAAG-----TCATAATATATTCGTTTAATGAGCT--------G--TCTCTGTATC-CA------CTGCAACTG--G-----TCTGATTCGTAT-------GTTT-TAT---TTATCCACTTTCAGATACCA---C-A-A--GACA------------------ACGCCGGGCCTGG-------GCGCCCA--------------CCT----AGAGAAGGAG-------AGGA------- | |
| droTak1 | scf7180000415310:752850-753104 + | TACTATCG--GAATTGGGCTGAACGTAA-AATGTCTCAATAG-----------------------------------ATTG-----------CCA-TAACTAG--T-------------------CAATGAGATTGGAAA-TGAGAATGCTGGCTGCC-----------------------GCCAAGGTTGTCTGGC----CATTA---TTAAAG-----TCATAATATATTCGTTTAATGAGCT---CC---G--TCTCTGTTTTGCA------CTGCAACTG--C-----TCTGATTCTTGTTT-----TTTT-TTTTATTTACTTACTTTCAGATACTACTGC-A-A--GACA------------------ACATCGGGCCTGT-------GCGCCCA--------------TCT----AGAGAAGGAG-------AGGG------- | |
| droEle1 | scf7180000491047:394805-395079 + | TACTATCG--GAATTGGGCTGCCAGTAA-AATGTCTCAATCG-----------------------------------GTGGCCATTGGGAAACCAGTAACCAG--TCTATGGAAATGGGAAT-GGGAATGGGAATGGAAA-TGAGAATGCTGGCTGCC-----------------------GCCAAGGTTGTCTGGC----CATTA---TAAAAG-----TCATAATATATTCTTTTAATGAGCT--GCC---G--TCTCTGTTTC-GA------ATGCAACTG--G-----TCTGATTCGT-------T-G-TT-TGT---TTATCTACTTTCAGATACCA---C-A-A--GTAA------------------ACATCGGGCCTGG-------GCGCCCA--------------ACT----GAAGAAGGAG-------AGGG------- | |
| droRho1 | scf7180000777865:25988-26244 - | TACTATCG--GAATTGGGCTGCCAGTAA-AATGTCTCAATCG-----------------------------------ATGGCCATTGGGAAACCAGTAACCAG--T-------------------CGATGGGAATGGAAA-TGAGAATGCTGGCTGCC-----------------------GCCAAGGTTGTCTGGC----CATTA---TAAAAG-----TCATAATATATTCGTTTAATGAGCT--GGC---G--TCTCTGTTTT-CA------ATGCAACTG--G-----TCTGATTCGT-------T-T-CT-TGT---TTACCTACTTTCAGATACCA---C-A-A--GTAA------------------ACATCGGGCCTGG-------GCGCCCA--------------ACT----AGAGAAGGAG-------AGGG------- | |
| droFic1 | scf7180000453912:1576858-1577143 - | TATAATCG--GAATTGGGCTGAGCGTAAAAATGTCTCAATTG-----------------------------------ATGGGAGTTGGGAAACCAGTAACAAG--TAAATCGAA---CGAACAGTCAATGGGATTGGAAATTGAGAATG-------CC-----------------------GCCAAGGTTGTCTGGC----TATAA---TAAAAG-----TCATAATATATTCGCCTAATCAGCTCTGCC---T--TCTCCGCTTG-CA------ATGCAACTT--G-----TCTGATTTGT-------TTTCAT-TTC---TTACCTACTTTCAGATACCA---C-A-A--TTCA------------------AAGTCGGGCCTGG-------GCGCCCA--A-----------TCT----TGTGAAGGAGAGACAG-GAGGAAGAAGA | |
| droKik1 | scf7180000302387:303774-303980 - | T--------------------------------------------------------------------------------------------------------------------------------------GAGAA-TGCGAATGCTGGCTGCC-----------------------GCCAAGGTTGTCTGGG----CAAAA---TAAAAG-----CCATAATATATTCATTTAATTAGCC--GCT---G--TCTCGTTTTA-CA------CCACAACTAGAG-----TCTGATTCGA-------TCTTTC-TTG---TTA--TTATTGCAGATACCG---C-A-A--GTGAACTAA-------------GCGCCGGGCCTTG-------GTGCCCCTTATCTGGAAGCTGTAGTAGGAGAGCAGGAG-------GAGG------- | |
| droAna3 | scaffold_13088:433531-433778 - | TGCTTTCG--GAATTGGGCTGAAAGT-------------------------------------------------------------GGAAACCAGTAA----------------------------------------------------------------------------------GCCAAGGTTGTCTGGT-GGGCATAATAATAAAAG-----CCACAATATATTCATTTAATTAGCT--TCTCAAGTCTCTCTATTCC-CG------CTGCAACTA--G-----TCTGATGTCCAATCTCTTTTTTT-TTT---TTATTTTATTTCAGAAAACT---TGGAA--GTTAACTGCGCAATCGCAG-CGCAGT---TGCAGGGCAACTTGCTTCCA--TTTTGGAAGTCATCA----AC--TGGGTG-------GGTG------- | |
| droBip1 | scf7180000396413:488796-489033 - | TGCTTTCG--GAATTGGGCTGA-------------------------------------------------------------------AAACCAGTAA----------------------------------------------------------------------------------GCCAAGGTTGTCTGGC----CACAA---TAAAAG-----CCATAATGTATTCATTTAATTAGCT--TCT---G--TCTCTTTTCT-CGCTATAACTGCAACTG--GCTATATCTGATATCCATAT-----CTCT-CTCTG-ATTTTTCCTTTCAGAAACTT---G-GAA--GTTAACTGCGCAATCGCAG-CGCAGT---TGCTGGGCAACTCGCTTCC----TTGGGAAGCCATCA----ACTGG--GAG----AG-CAAC------- | |
| dp5 | 2:30708822-30709120 + | CAGACTGA--GAATTGGGGTC--------------TCAATAATTTACGAGTTGGAGCAATTTCCATTTCCAATGGAAATGCCAGTTGGTAGACCAGTAACCAG--T-----------------------GTCAGTGCCAG-TGCCAGTGCCCCCCGCTCGGTGGCCTGTGCTATGTGGTGGTCCAAGGCTGTCTGGCTGGGCATAA---TAAAAAACCGATGATAATGTATGCGTTTAATTAGCT--CCA---C-----------------------------------------------CATTC-------TC-TTT---CTCTCTCTTTTTAGAGATCG---T-AAAGTGTGAACTGCGCAGTCGCAGTCGCAGCAGCGCCTGT-------GCGCCGG--------------CGC----ACAGCCGGAG-------AAGC------- | |
| droPer2 | scaffold_6:6108504-6108793 + | CAGACTGA--GAATTGGGGTC--------------TCAATAATTTACGAGTTGGAGCAATTTCCATTTCCAATGGAAATGCCAGTTGGTAGACCAGTAACCAG--T-----------------------GTCAGTGCCAG-TGCCAGTGCTCCCCGCTCGGTGGCCTGTGCTATGTGGTGGTCCAAGGCTGTCTGGCTGGGCATAA---TAAAAAACCGATGATAATGTATGCGTTTAATTAGCT--CCA---C-----------------------------------------------CATTC-------TC-TTT---CTCTCTCTTTTTAGACATCG---T-AAAGTGTGAACTGCGCAGTCGCAGTCGCA---------GT-------GCGCCGG--------------CGC----ACAGCCGGAG-------AAGC------- | |
| droVir3 | scaffold_12875:13354579-13354624 + | GAA---------------TTAAAAGTAA-AAGGCTT----------------------------------------------AATTGGAAAAACAATAAATAG--T-------------------TAAT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droMoj3 | scaffold_6328:2342026-2342051 + | GG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGCACA--------------GCA----ACAG-------------CAGCGAGGCCG | |
| droGri2 | scaffold_15252:14884553-14884558 + | AGCAGC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/19/2015 at 03:18 PM