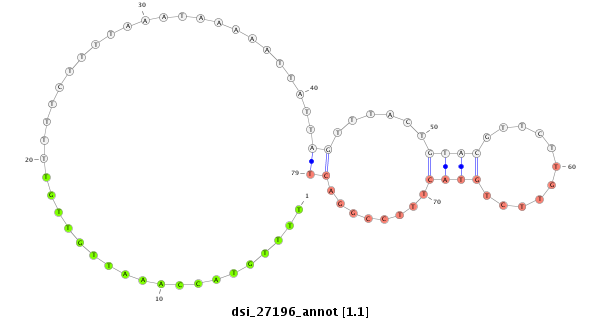
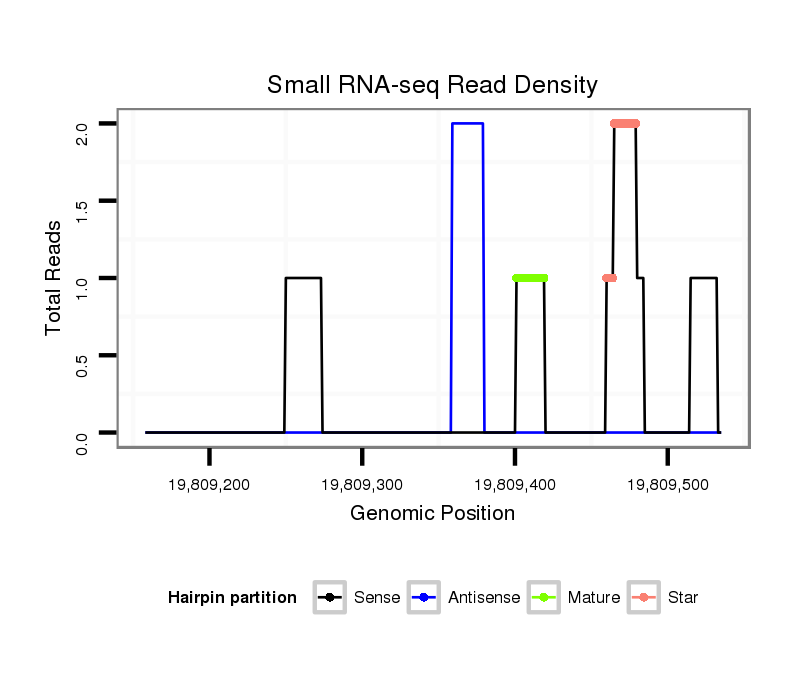
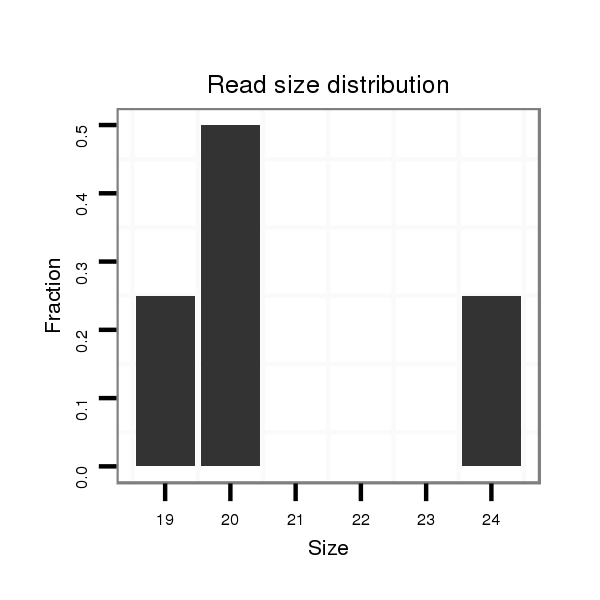
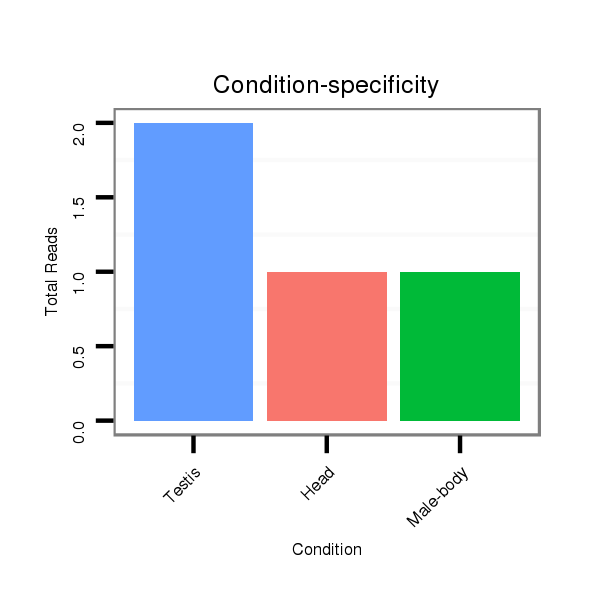
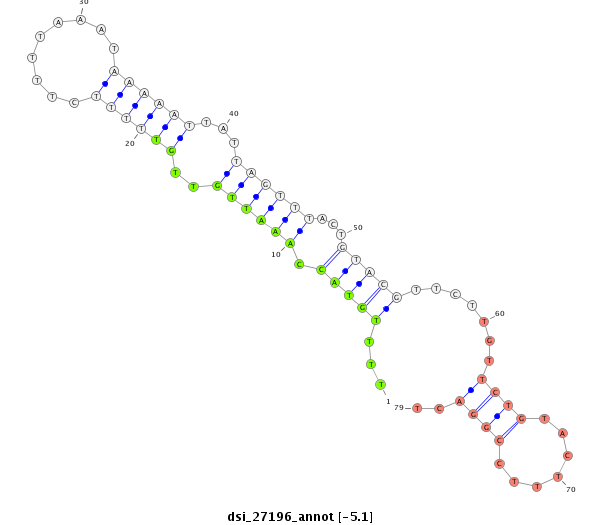
ID:dsi_27196 |
Coordinate:2l:19809208-19809485 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -5.1 | -4.9 | -4.8 | -4.7 |
 |
 |
 |
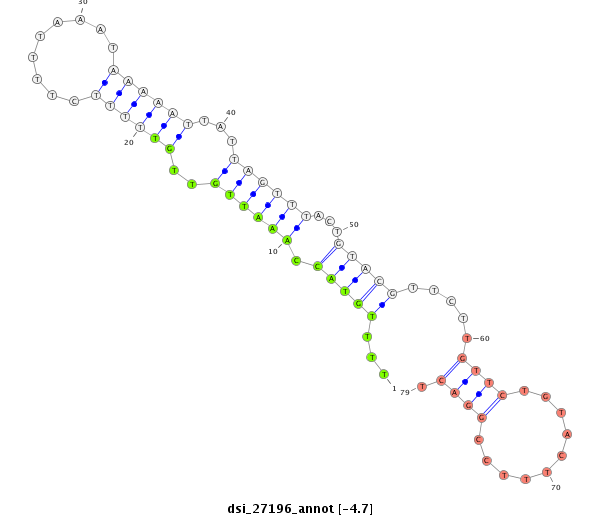 |
intergenic
No Repeatable elements found
|
CCCACGCCATGATCTTGGAGTTCATCAGGTCACGGCCGCCGCTGAAGAAGGTGTGTAAAGCCCCCAATGCCAGTTCCCCAATGAATTCCCAATCCATTCGCACGGCCAGTACCCAAGTCAACCGCAATCCCGACAACAATCCCTCCTCATCAAAATCCCCATTTGCCTCTGGCAGAATGTGAAATATTGATGCAAAGGCTTTCCCTTCTTCCGACTTCATTTCCTTTGCCATGATTCCTCATTTTTTGTACCAAATTGTTGTTTTTCTTTTAAATAAAAATTATTAGTTTACTGTACGTTCTTGTTCTGTACTTTCCGGACTCATCAGGCCAGCGATCGACAATTGGGCCCACCGCGCATGTGCGAGCCTTCCCCGCG
***************************************************************************************************************************************************************************************************************************************************..........................................((......((((...........))))........))******************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | O001 Testis |
O002 Head |
M024 male body |
SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR553485 Chicharo_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................................................................................TTTTGTACCAAATTGTTGT.................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................TGTTCTGTACTTTCCGGACT........................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................TGTACTTTCCGGACTCATCA................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................CATGTGCGAGCCTTCCCC... | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................TCCATTCGCACGGCCAGTACCCAA...................................................................................................................................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................AACGTGAAATATCGAGGCAAAG..................................................................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...GCGCCATGATCTCGGTGTTCA.................................................................................................................................................................................................................................................................................................................................................................. | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................TACTGTCTGTTCTTGTTGTGT.................................................................... | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................CTAACCGCAATCCCGACGA.................................................................................................................................................................................................................................................. | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................GTCTCTACTTTCGGGACTC....................................................... | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................CCGCCGCTGAAGACG........................................................................................................................................................................................................................................................................................................................................ | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................................................TGTTGTTGTTATTTTAAAAAAA.................................................................................................... | 22 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GGGTGCGGTACTAGAACCTCAAGTAGTCCAGTGCCGGCGGCGACTTCTTCCACACATTTCGGGGGTTACGGTCAAGGGGTTACTTAAGGGTTAGGTAAGCGTGCCGGTCATGGGTTCAGTTGGCGTTAGGGCTGTTGTTAGGGAGGAGTAGTTTTAGGGGTAAACGGAGACCGTCTTACACTTTATAACTACGTTTCCGAAAGGGAAGAAGGCTGAAGTAAAGGAAACGGTACTAAGGAGTAAAAAACATGGTTTAACAACAAAAAGAAAATTTATTTTTAATAATCAAATGACATGCAAGAACAAGACATGAAAGGCCTGAGTAGTCCGGTCGCTAGCTGTTAACCCGGGTGGCGCGTACACGCTCGGAAGGGGCGC
********************************************************..........................................((......((((...........))))........))*************************************************************************************************************************************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
SRR902009 testis |
SRR553485 Chicharo_3 day-old ovaries |
M053 female body |
SRR618934 dsim w501 ovaries |
M023 head |
M024 male body |
GSM343915 embryo |
M025 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................................GGAACAAGGCTGAAGT............................................................................................................................................................... | 16 | 1 | 2 | 2.00 | 4 | 0 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................AGGGAAGAAGGCTGAAGTAAA............................................................................................................................................................ | 21 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................AGGGTGCCGGGCATGGGTTT..................................................................................................................................................................................................................................................................... | 20 | 3 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................ATGGAACAAGGCTGAAGT............................................................................................................................................................... | 18 | 2 | 4 | 1.50 | 6 | 1 | 3 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................GGAACAAGGCTGAAG................................................................................................................................................................ | 15 | 1 | 6 | 1.33 | 8 | 2 | 1 | 2 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................ATGGAACAAGGCTGAAGTG.............................................................................................................................................................. | 19 | 3 | 12 | 1.25 | 15 | 4 | 4 | 1 | 0 | 1 | 3 | 0 | 0 | 2 | 0 | 0 |
| .......................................................................................................................................................................AGTCCGTCTTACACGTTAT................................................................................................................................................................................................ | 19 | 2 | 5 | 1.20 | 6 | 1 | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................GAACAAGGCTGAAGT............................................................................................................................................................... | 15 | 1 | 7 | 0.71 | 5 | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................CATTTCTGGGGTTAAGGTC................................................................................................................................................................................................................................................................................................................. | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................TGACGTTAGGCCTGATGTTAG............................................................................................................................................................................................................................................. | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................TGGAACAAGGCTGAAGTG.............................................................................................................................................................. | 18 | 3 | 20 | 0.40 | 8 | 2 | 3 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................GGAACAAGGCTGAAGTG.............................................................................................................................................................. | 17 | 2 | 10 | 0.40 | 4 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................AATGGAACAAGGCTGAAGT............................................................................................................................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................AGTTAGGGAGGAGGAGATTT............................................................................................................................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................TACCAGGGAGTAAAACACAT................................................................................................................................ | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................GATGGAACAAGGCTGAAGT............................................................................................................................................................... | 19 | 3 | 11 | 0.27 | 3 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................TTAAGGGTTAGGTATGCCGG................................................................................................................................................................................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................AGGGTGCCGGGCATGGG........................................................................................................................................................................................................................................................................ | 17 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............GAATGTCAAGTAGTCCAGT.......................................................................................................................................................................................................................................................................................................................................................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................TGGGTTCAGATGGTGTTATG........................................................................................................................................................................................................................................................ | 20 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................TGGAACAAGGCTGAAGT............................................................................................................................................................... | 17 | 2 | 13 | 0.23 | 3 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................ATGGAACAATGCTGAAGT............................................................................................................................................................... | 18 | 3 | 20 | 0.20 | 4 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................GAAAGGGGAGAAGCCGGAAG................................................................................................................................................................ | 20 | 3 | 19 | 0.16 | 3 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| ............AAAATGTCAAGTAGTCCAGT.......................................................................................................................................................................................................................................................................................................................................................... | 20 | 3 | 15 | 0.13 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................................AGAGTTAGGTAAGCG..................................................................................................................................................................................................................................................................................... | 15 | 1 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................GGCGGAGTGGTTTGAGGGGT......................................................................................................................................................................................................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................AGTCCGACTTACACGTTAT................................................................................................................................................................................................ | 19 | 3 | 20 | 0.10 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................................................................................................................................................................GAAGGCTGAAGCAAA............................................................................................................................................................ | 15 | 1 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................TTCGTTAAGGGTTAGGTA......................................................................................................................................................................................................................................................................................... | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................................................................................................AAGGAATTGGTACTAAG............................................................................................................................................. | 17 | 2 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................TCAAGTAGTCCAGTTCT....................................................................................................................................................................................................................................................................................................................................................... | 17 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................AGGGAACAATGCTGAAGTG.............................................................................................................................................................. | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................TTGAGAGTTAGGTAAGC...................................................................................................................................................................................................................................................................................... | 17 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................GGGTTACCTAATCGTTAGGT.......................................................................................................................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................AGTCCAGTGCTGGGGCCG................................................................................................................................................................................................................................................................................................................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................GAGAGTTAGGTAAGCG..................................................................................................................................................................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................GAAAGGGGAGAAGCCGGAA................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................TTTTAGGGGTAGAACGAGAC............................................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................CCGTCATGGATTCAGTTA................................................................................................................................................................................................................................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 2l:19809158-19809535 + | dsi_27196 | CCCACGCCATGATCTTGGAGTTCATCAGGTCACGGCCGCCGCTGAAGAAGGTGTGT-AAAG---------CCC------------------------------------------------------CCAAT------------------------GC---CAGTTCCCCAATGAATTCCCAATCCAT---TCGCA----------------CGGCCAGTACCCAAGT------CAACCGCAATCCCGACAACAATC---------C-----------------CTCCTCATCAAAAT------------------------------------------CCC----------------------------------------CATTTGCCTCT-----------GGCAGAATGTGAAA--------TATTGATGC-AAAGGCTTTC-CCT-TCTTCCGACTTCATTTC--CTTTGCCA-TGATT-----------------------------------------------------------------------------CCT-----------CA-TTT----TTTGTACC-------------AAATTGTTGT-TTTTCTTTTAAATAAAAATTATTAGTTT----------ACTGT------------A--------------------------------------------CGTTCTTG------------------T--------------------------------------------------TCTG---------------TACTTT-C---CGGACTC---------------ATCAGGCCAGCGATCGACAATTGGGCCCACC---GCGCATGTGCGAGCCTTCCCCGCG |
| droSec2 | scaffold_18:233290-233668 + | CCCACGCCATGATCTTGGAGTTCATCAGGTCACGGCCGCCGCTGAAGAAGGTGTGT-AAAG---------CCC------------------------------------------------------CCAAT------------------------GC---CAGTTCCCCAATGAATTCCCAATCCAT---TCGCA----------------CGGCCAGTACCCAAGT------CAACCGCAATCCCGACAACAATC---------C-----------------CTCCTCATCAAAAT------------------------------------------CCC----------------------------------------CATTTGCCTCT-----------GGCAGAATGTGAAA--------TATTGATGC-AAAGGCTTTC-CCT-TCTTCCGACTTCATTTG--ATTGGCCG-TGATT-----------------------------------------------------------------------------CCT-----------CA-TTT----TTTCTACC-------------AAATTGTTGT-TTTTCTTTGAAATAAAAATTATTAGTTT----------ACTGT------------A--------------------------------------------CGTTCTTG------------------TT-------------------------------------------------TCTG---------------TACTTT-C---CGGACTC---------------ATCAGGCCAGCGATCGCCAATTGGGCCCACC---GCGCATGTGCGAGCCTTCTCCGCG | |
| dm3 | chr2L:20339644-20340029 + | CCCACGCCATGATCTTGGAGTTCATCAGGTCACGGCCGCCGCTGAAGAAGGTGTGT-AAAG---------CCC------------------------------------------------------CCAAT------------------------GC---AAGTTCCCCAACGAATTCCCAATCCAC---TCGCA----------------TGGCCAGTACCCAAAT------CAACCGCAATCCCAACTGCAATC---------C-----------------CTCCTCATCAAAAT------------------------------------------CCC----------------------------------------CATTTCACTTT-----------GGCAAAGTGTGAAA--------TATTAAACT-ACAGGTTTTC-CCT-TTTTCCGACTTCATTTC--ATTCGCCG-TGATT-----------------------------------------------------------------------------CCT-----------AATTTT----TTTGTACC-------------AAATTGTTGT-TTTTCTTCGAAATAAAAATTATTAGTTT----------ACTGT------------A--------------------------------------------CGTTCTTGTTTC------------TGTT-------------------------------------------------TCTG---------------TACTTT-C---CGGACCC---------------ATCAGGCCAGCGATCGCCAATTGGGCCCGCC---GCGCATGTGCGAGCCTTCTCCGCG | |
| droEre2 | scaffold_4845:4084294-4084680 - | CCCACGCCATGATCTTGGAGTTCATCAGGTCACGGCCGCCGCTGAAGAAGGTGTGT-AAAG---------CCC------------------------------------------------------CCAGT------------------------CC---CAGTGCCCCAACCCATTCCCAAATCAT---CCGCA----------------CGGCCAGTACCCAAAT------CAAAA------------GCAATT---------C-----------------CCCATCATCAAAAT------------------------------------------CCC----------------------------------------CATTCGCCGCT-----------TGCAGAGTGTGATG-----AAGTATTGATCT-TGAGGCTTTC-CC----TTCCAACTTCATTTC--ATTGGCCT-TGATT-----------------------------------------------------------------------------CCCCTCCCCACTGACA-TTT----TTTGTAGCAAATG---C-ATCAAATTGTTGT-TCTCCTTTGAAATAAAAATTATTAGTTC----------ACTGT------------C--------------------------------------------CGTTTTCA------------------TT-------------------------------------------------TCTG---------------TATTTC-C---CGGACCC---------------ACCAGGCCAGCGATCGCCAACTGGGCCCGCC---GCGCATGTGCGAGCCTTCTCCGCG | |
| droYak3 | 2R:6711570-6711966 + | CCCACGCCATGATCTTGGAGTTCATCAGGTCACGGCCGCCGCTGAAAAAGGTGTGT-AAAG---------CCC------------------------------------------------------CCAGT------------------------CC---CAGTGCGCCAGGGAATTCCCAATCCAA---CCGCA----------------CGGCCAGTACCCAATC------CAACCGCAATCCCAACTGCAATC---------C-----------------CTCATCATCAAAAT------------------------------------------CCC----------------------------------------CATTTGCCACT-----------TGCAGACTGTGAAATATTGAAATATTGATCT-TGAGGCTTTC-CCA-CCTTTCGACTTCATTTC-ATTTTGCCG-TGATT-----------------------------------------------------------------------------CCC-----------CA-TTT----TTTGTACCAAATG---C-ATCAAATTGTTGT-TCTCCTTTGAAATAAAAATTATTAGTTC----------ACTGT------------C--------------------------------------------CGTTTTTA------------------TT-------------------------------------------------TTTG---------------TATTTT-C---CGGACCC---------------ACCAGGCCAGCGATCGCCAATTGGGCCCGCC---GCGCATGTGTGAACCTTCTCCGCG | |
| droEug1 | scf7180000409767:947154-947563 - | CCCACGCCATGATCTTGGAGTTCATCAGGTCACGGCCGCCGCTGAAGAAGGTGTGT-AGCG---------CCC------------------------------------------------------CCAAT------------------------CC---CATTGCCTCAACAAATTCCAAATCCAT---CCGCA----------------TCGCAAGTCCCAAAAG------CAACCTCAATTCCCACTGCAATACCACTG---C-----------------CCCATCATCAAAAT------------------------------------------CCC----------------------------------------CATTTGCCGCT----------TTGCGGAGTGTGAAA--------TATTGATCT-TGTGGCTCTT-ATT-CCTGACCATTTCATTTC-TGTTTGTCT-TGATCTCATGACCTT-----------------------------------------------------------------TGA-T-----------TG-TTT----TTTGTACCAAAAA---AAAGCAAATTACTTT-TCGCTCTTCGAATAAAAATTATAAGTTC----------ACTAT------------T--------------------------------------------TAATATTG------------------TATC-----------------------------------------------TTTG---------------TGATTT-C---CGGATCC---------------TTCAGGCCAGCGATCGCCAATTGGGGCCACC---CCGCATGTGCGAACCTTCTCCGCG | |
| droBia1 | scf7180000302422:8025175-8025580 - | CCCACGCCATGATCTTGGAGTTCATCAGGTCACGGCCGCCGCTGAAGAAGGTGTGT-AAAG---------CCC------------------------------------------------------CCGAT------------CCCAATTCCAATCC---AACTGCCACAACAAATTCCAAATCCAT---GCGCA----------------CCGCCAGTCGCAAAGG------CGCCCTCAATTCCAACTGCAGTC---------C-----------------CCCATCACCAAAAT------------------------------------------CCC----------------------------------------CATTTGCCGCT-----------TGCAGAGTGTGAAA--------TATTGATCC-GGTGGCTTTC-CTT-CCCGCCGATTCCATTTC--ATTTGTCC-TGATCACTTGA-------------------------------------------------------------------------------------CC-TTT----TTTGTACCCAAAGAAAA-ATCCAATTGTTGT-TCGCCTTTCGAATAAAAATTATTAGTTT----------ACTAT------------A--------------------------------------------CGTTTCTG------------------TA-------------------------------------------------TCTG---------------TACTTC-T---CGGATCC---------------ACCAGGCCAGCGAGCGCCAACTGGGGCCGCC---GCGCATCTGTGAGCCTTCTCCGCG | |
| droTak1 | scf7180000415115:228486-228918 - | CCCACGCCATGATCTTGGAGTTCATCAGGTCACGGCCGCCGCTGAAGAAGGTGTGT-AAAG---------CCC------------------------------------------------------CCGAT-----TCCCATTC-CAATTGCAATCC---CACTGCCACAACCAATTCCCAATCCAT---CCGCA----------------CCGCCAGTCCAAAAGCCAAATGCAACCTCAATTCGAACTGCAGTCACAGTC---A-----------------CCCATCATCAAAAT------------------------------------------CCC----------------------------------------CATTTGCCGCT-----------TGCAGAGTGTGAAA--------TATTGATCT-TGTGGCTTTC-CTT-CCTGTCGATTCCATTTCATGTTTGTCTGTAATCGCATAACC----------------------------------------------------------------------CT------------C-ATT----TTTGTACCAAAAA---A-ATTCAATTGTTGT-TCTCCATTCAAATAAAAATTATTAGTTT----------ACTAT------------A--------------------------------------------CTTTTCTGTATC------------TGTA-------------------------------------------------TCTG---------------TACTTT-T---TGGAACC---------------ACCAGGCCAGCGAACGTCAGCTGGGGCCACC---GCGCATCTGCGAGCCTTCTCCGCG | |
| droEle1 | scf7180000491186:298369-298769 + | CCCACGCCATGATCTTGGAGTTCATCAGGTCACGGCCGCCGCTGAAGAAGGTGTGT-AAGA---------CCC------------------------------------------------------TCAAT------------------------CC---CAGTGCCACGGCAAATTCCAGATCCAT---CTGCA----------------CCGCCAGTTCCGAAAG------CAACCTCAATTCCAACTGCAGTC---------C-----------------CCCACCATAAAAAT------------------------------------------CCC----------------------------------------CATTTGCCGCT-----------TGCACAGTGTGAAA--------TATTGATCA-TGTGGCTTTC-CTT-CCTGTCGATTCAATTTC--ATTTGTCT-TGATCTCGTGACCAT-----------------------------------------------------------------TTCTT-----------CT-TTT----TTTGTACCAAAAG---A-ATCAAATTGTTTT-TCTTACTCGAAATAAAAATTATTGGTTC----------ACCTT------------T--------------------------------------------CGTTTCTG------------------TA-------------------------------------------------TCTG---------------TATTTTCC---CGGATCC---------------AACAGGCAAGTGAACGCCAATTGGGACCGCC---GCGCATGTGCGAACCGTCGCCGCG | |
| droRho1 | scf7180000774359:17448-17850 - | CCCACGCCATGATCTTGGAGTTCATTAGGTCACGGCCGCCGCTGAAGAAGGTGTGT-AAAG---------CCC------------------------------------------------------CCAAT------------------------CC---CACTGCCACAACGAATTCCAAATTCAT---CCGCA----------------CCGCCAGTGCCGAAAG------CAACCTCAATTCCAACTGCAATC---------C-----------------CCCATCATCGAAAT------------------------------------------CCC----------------------------------------CATTTGCCGCT-----------TGCAGAGTGTGAAA--------TATTGATCT-CGTGGCTTTC-CTT-CCTGTCGCTTCCATTTT--GTTTGTCT-TGATCTCTTCACCAT-----------------------------------------------------------------T---------------TT-CTT----TTTGTACCGAAAG---A-ATCAAATTGTTTT-TCTCATTTAAAATAAAAATTATAAGTTC----------ACTAT------------C--------------------------------------------CGTTTCTGTTTC------------TGCA-------------------------------------------------TCTG---------------TATTTTCC---CGGAACC---------------AACAGGCCAGTGAACGCCAATTGGGGCCGCC---GCGCATGTGCGAACCTTCGCCGCG | |
| droFic1 | scf7180000454082:732493-732890 + | CCCACGCGATGATCTTGGAGTTCATCAGGTCACGGCCGCCGCTAAAGAAGGTGTGTAGTAGA-------GCCC------------------------------------------------------CCAAT------------------------CC---CAGTGCCACAGCCAATTCCAAATCCAT---CCGCA----------------TCGCCAGTCCCAAAAT------CAACCTCAATTCCAACTGCAATC---------G-----------------GCCATCATCAAAAT------------------------------------------CCC----------------------------------------CATTTGCCGCT-----------TGTAGAGTGTGAAA--------TATTGATCT-TGTGGCTTTC--TT-CCTGCCGATTCCATTTC--ATTTGTCT-TGATCTCTTA-C---A--------------------------------------------------------------------C-----------TG-TTT----TTTGTACCGCATG---A-ATCAAATTGTTTTTTCTCCGTTCAAATAAAAATTGTAAGTTT----------ACTAT------------C--------------------------------------------CGTTTTC-----------------TTTA-------------------------------------------------TCTG---------------TGCTTTTT---CGGATCC---------------ATCAGGCCAGCGATCGCCAGTTGGGACCGCC---GCGGATGTGCGAACCTTCGCCGCG | |
| droKik1 | scf7180000302389:270410-270767 - | CCCATGCCATGATCTTGGAGTTCATTAGGTCACGACCGCCGCTCAAGAAGGTGTGT-AAAG---------CTC------------------------------------------------------CCAAA------------CCCCCT------GC---TAAACCAACAACAAATTCCAAGTCAAA---GTATA----------------TTCTCAATA------T------GAATCTCAATTTGAAGTCCAAT-----------------------------TCAATATCAAAAT------------------------------------------CCC----------------------------------------CACTTGGCGCT-----------TGCAGTGTGTGAAA--------TTTTAAAC--------------------GTTGATTTAATTTC--CCTTGACT-TGCCTTC------------------------------------------------------------------------------------------------------------------------------------TTGCTTTCAAATAAAAATTGTTAGTTTTTAATAGAACACTGT------------C--------------------------------------------TAACCTTG------------------AGTG----------------------------------------------TTT-----------------TGTTTT-T---CGAATTT--GTATTTAAATTTTGACAGGCCTCGGAACGGCAATTAGGACCGCC---GCGCATGTGCACACCCTCGCCCCG | |
| droAna3 | scaffold_12916:7517193-7517631 - | CCCATGCCATGATCTTGGAGTTCATCAGGTCACGGCCGCCACTCAGAAAGGTGTGT-ACGACCACCAAAGCCACCACC---------------------------------------------------AATCAGAACCCCATTCCCAATCCCAATCC---CACTGCCACATCAAATTCAAAATCAAGTAACTGCAACCAGTCAAGT---AT-------CATCCAAAT------CAAAATCAAAACCAACTGCAAAC---------C-----------------AAATCAATCAAAAT------------------------------------------CTC----------------------------------------CATATGCCGCT-----------TGCAGAGTGTGAAA--------TTTTTACGACTTCGACTTTC-ATT-C--GACGATTCCATTTG--GTTTGTCT-TGACCTCGCG----TTA-------------------------------------------------------------------------------TT-TTT----TTTGTACCGAATC---C-GTAAAATTGTTTC-T-TGTTTTCAAATAAAAATTACTAGTTTCTCTTGCAAAAATCTCAAAAAAAATGTA--------------------------------------------TGTTTTTC---------------------------------------------------------------------------------------------------------T---------------TAAAGGCCTCTGAGCGGCAGTTAGGACCGCC---TCGTATGTGCACACCCTCGCCCCG | |
| droBip1 | scf7180000396554:133717-134149 - | CCCATGCCATGATCTTGGAGTTCATCAGGTCGCGGCCTCCGCTCAGGAAGGTGTGT-ACCAC---TAAAGCCA------------------------------------------------------CCAATCAGAACCCCATTCCCAATCCCAATCC---CACTGCCACATCATATTCAAAATCAAGTAACTGCAACCAGTCAAGTACTAT-------CGTCCAAAT------CACAATCAAGACCAACTGCCAGC---------CACGG--------------AATCCATCAAAAT------------------------------------------CCC----------------------------------------CCTATGCCGCT-----------TGCAGAGCGTGAAT--------TTTTTACCC-AGCGACTTTC-CTT-C--GACGAACC-CTATG--ATTTGACT-TGACCTCACG----TTA--------------------------------------------------------------------------------T-TTT----TTTGTAACGAATC---C-ATCAAATTGTTTC-T-TGTTTTGAAATAAAAATTACTAGTTTCACTTGCAAAGATCT-AAACAAAAT-TA--------------------------------------------TGTTTTC----------------------------------------------------------------------------------------------------------T---------------TAAAGGCTTCTGAGCGGCAGTTAGGACCGCC---GCGGATGTGCGAACCCTCGCCCCG | |
| dp5 | 4_group4:2583881-2584224 + | CCCATGCCATGATCTTGGAGTTCATTAGGTCACGGCCGCCATTGAAGAAGGTGTGT-AGTG---------CCC------------------------------------------------------CCAAT------------------------CCAACCACAACCACAACCATTTCCAGTCCCAA---GTGAA----------------------------------------------------------------------------------------------------CC------------------------------------------CCC----------------------------------------CCTCTGG-GCT----------TTCTGGCGGGT--------------------------TT----AT--AT--T----------------------T-TTACTTCACT----TTC--------------------------------------------------------------------------------T-TTA----TGGAGGAAGCATC---G-CTGCATATATTTT-G-TGGCTTCCAATAAAATTTAACAGTTCCATTTTCGAAG----CAAAACTAATTTTCGTTGCCATATTTTTTCTTTTCTTGTTCTATTTCCCTT------------------------------------------------------------------------------------CCC---------------CACCTT-C---ATCGTCC---------------ACCAGGCCTCCGAACGGCAACTGGGCCCGCC---CCGGATGTGCACGCCCACGCCCAG | |
| droPer2 | scaffold_10:1593355-1593697 + | CTCATGCCATGATCTTGGAGTTCATTAGGTCACGGCCGCCATTGAAGAAGGTGTGT-AGTG---------CCC------------------------------------------------------CCAAT------------------------CCAACCACAACCACAACCATTTCCAGTCCCAA---GTGAA-----------------------------------------------------------------------------------------------------C------------------------------------------CCC----------------------------------------CCTCTGG-GCT----------TTCTGGCGGGT--------------------------TT----AC--AT--T----------------------T-TCACTTCACT----TTC--------------------------------------------------------------------------------T-TTA----TGGAGGAAGCATC---G-CTGCATATATTTT-G-TGGCTTCCAATAAAATTTAACAGTTCCATTTTCGAAG----CAAAACTAATTTTCGTTGCCATATTTTTTCTTTTCTTGTTCTATTTCCCTT------------------------------------------------------------------------------------CCC---------------CACCTT-C---ATCGTCC---------------ACCAGGCCTCCGAACGGCAACTGGGCCCGCC---CCGGATGTGCACGCCCACGCCCAG | |
| droWil2 | scf2_1100000004577:1974558-1975052 + | dwi_1574 | CCCATGCCATGATATTGGAGTTCATCAGGTCACGGCCGCCATTGAAGAAGGTGGGT-AGCAGCAGCATAAGCAGCAGCAGCAACGGCAATGGCAGTAAAGTCGAAACGCCGACAACTACTACTGGCCCCAAT------------------------CT-----------------A-ACCAAATCCAA---AGGCA----------------TAACCGCAATCCAAAT------GAATCCAAGTCCCAATAACAA----------------------------------------AATCGAGTATCCAATTTAACTTTCCCTTCTTCATCTTCCCTCCCACTCACTCTTCCACTCTTTGTCACACTCACACACCATCTACCTTACTCTGTCGCTCTTCACATCTCTGCTGTGTGTT----------------------------------------------------------CGTCA-AGAAAATTTAA---T--TTTGTTTTAATAAAAATTGAAATTGTTGTCTGTAACTCATTTTTGCATTACAAATTGTTGTTG-------------------------------------------------------AT-TTACCT---------------------------------TTGT------------T--------------------------------------------TG---------------------------------------------------------------------------TTTG---------------TTTTTT-TGTTTGGAAAATAATATTT--ATATATATAGGCCTCGGAGCGTCAATTGGGGCCGCCATTGTTGAAGTGCATCCCATCGCCGAG |
| droVir3 | scaffold_12963:11849133-11849516 - | dvi_11414 | CCCACGCGATGATCTTGGAGTTCATTAGGTCACGGCCGCCATTAAAGAAGGTGTGT-GTTGC-------GTTC------------------------------------------------------CCTAT-----------GCCCATT------TC---GAATAGCCAAACAAATGCTTAATATGA---CTGTC----------------CAAT----GTTCCAAT------GTTCA---------ACTA--GTTACATATATGCACTGTCT----------CC---------TAT------------------------------------------CGC----------------------------------------GCTCTTTCGCT-----------TTCAATTTGTGAAT--------TTTTGTGTT-A---TT----AT--GC-----------ATATACTATATGTATATATACATATATAT----------------------------------------------------------------------AT------------A-TAT------TTTGCTAAAAT---A-AACCCATTCGTTT-ACTCAATTCAATTAA-------------------------------------------------------------------------------------------------TGGCATATGGA-------------------------------------------TTTTCATTAATGTTTACAACAAATGCTCCTCATTGT-T---TTC--TT---------------TAAAGGCCTCGGAACGCCAACTGGGCCCACC---ATTGAAATGCACGCCCTCGCCCCG |
| droMoj3 | scaffold_6500:20549146-20549559 - | CGCATGCGATGATCTTGGAGTTCATCCGGTCACGGCCGCCATTAAAGAAGGTGTGT-GTTGC-------GTTC------------------------------------------------------CCAAT-----TTCCA------TA------GC---AAATACTTCAGCAAATGCCTAATGGAG---TCGTT----------------CAAGACCATGCAAAAT------CTAT------------TGATATT---------TATTGTCTCTTTCAAATCCACCTC-----TCT------------------------------------------CTC----------------------------------------ACTCTCACTCT-----------ATCAAAATGTGAAC--------AATTTTTGC-TGTTGTAT------GTATGAAGATGTTAATAT--ATAT----------ATAT----------------------------------------------------------------------------------------A-TATTTTTTATAAAATAAATA---C-ATCGG----------------------------------------------------------------------------------------------------CTTGCTCACTTTTAACATAC-TTGCATTTGAGT-----AATAAATGTAAAAACTATTATATATAGCAACAATTAATTATTATTAATTATG---------------AATTGC-T---CTAATT-----------------CAAGGCCTCGGAACGCCAGCTAGGTCCCCC---TCTGAAGTGCACGCCCTCACCCCG | |
| droGri2 | scaffold_15126:5402227-5402462 - | CCCATGCGATGATCTTGGAGTTCATCAGGTCACGGCCGCCATTAAAGAAGGTGTGT-GTTGC-------GTCC------------------------------------------------------CCAAT-----TCCCATACCCACT------AC---AGAAACCAAAACGAATACCCA------------------------------------------------------------------AC----------------------------------------------AT------------------------------------------CTC----------------------------------------TATT----------------------GACT-----------------------------------------------------ATT--ATTTACCA-TTATT-----------------------------------------------------------------------------TAT--------TACTA-TTT----TTTATTACAAA-----------------------------------------------------------TATTT-ATT--ACTT-TT--------------------------------------------CTTTTGT----------------------------------------------------------------------------------------------------------G---------------TTCAGGCCTCGGAACGTCAGTTGGGCCCACC---GCTCAAGTGTACGCCTTCACCCAG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/19/2015 at 03:11 AM