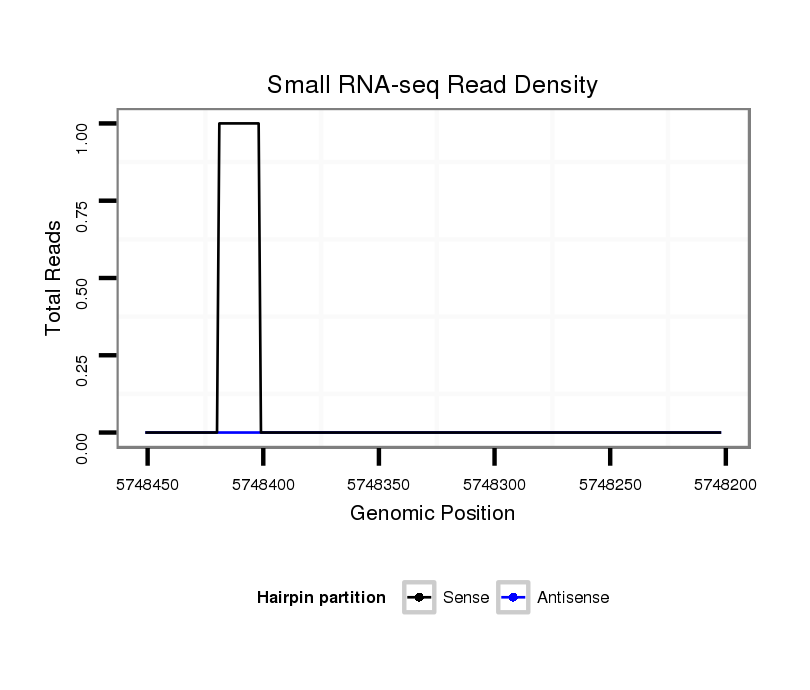
ID:dsi_27082 |
Coordinate:2r:5748252-5748401 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
exon [2r_5747949_5748251_-]; CDS [2r_5747949_5748251_-]; intron [2r_5748252_5753713_-]
No Repeatable elements found
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TGCGTATACGCAACATTGGCCGTCGGGAGTAGGTGGGTGGTGTGTGGAGGGCACTGGAAGGCGGGTCCTGTTGGTTTTGCTTTTGATGTAAAGAGGCACTCGAGGCAATTTTACGCTCTTATCCCAATCAACGAGCAGTAATGGCGAGCAGCACAAGGGCGACCGATAGCTAACCTTTGTTGTACGTTACACCGTCCTAGAACAACCATCAGACACATCAGCAACAGGGCCAGCCGCAACAGCCACATCA **************************************************...((((..(((((..((....))..(((((((.(((((......))).)))))))))..((((((((..............)))).))))(((((.((((((.((((((........))...)))))))))).)))))..)))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
O001 Testis |
SRR553487 NRT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
M025 embryo |
M024 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................CGACCGATAGTTAACC........................................................................... | 16 | 1 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................GTGGGTGGTGTGTGGAGG........................................................................................................................................................................................................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................GACCGATAGCGAACC........................................................................... | 15 | 1 | 3 | 1.00 | 3 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................GCGACCGATAGTGAACC........................................................................... | 17 | 2 | 6 | 0.67 | 4 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................CGACCGATAGCGAACC........................................................................... | 16 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................CGAGACCGATAGCTAAC............................................................................ | 17 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................CGACCGATAGTGAACC........................................................................... | 16 | 2 | 20 | 0.30 | 6 | 1 | 4 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................CGACCGATAGCGAAC............................................................................ | 15 | 1 | 8 | 0.25 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................TAGGCAGTAGGTGGCTGGTGT............................................................................................................................................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................CGACCGATAGTTAACCA.......................................................................... | 17 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................GCGACCGATAGCGAA............................................................................. | 15 | 1 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................TGCGACCGATAGCTAACTA.......................................................................... | 19 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................CATCATCATCAGGGCCAGGCG.............. | 21 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............ATTTGCCGTTGGGGGTAGG......................................................................................................................................................................................................................... | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................CGTCGAGAGTAGTTGGGGG................................................................................................................................................................................................................... | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................................................................................AGTAATCGCGATCAGCA................................................................................................. | 17 | 2 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................CGACCGATAGTTAACT........................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................ACCGATAGCTATCCTG......................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............ACGTTGGCCTTCGGGA.............................................................................................................................................................................................................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................AAGGGGTACTCGAGGCTA.............................................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
ACGCATATGCGTTGTAACCGGCAGCCCTCATCCACCCACCACACACCTCCCGTGACCTTCCGCCCAGGACAACCAAAACGAAAACTACATTTCTCCGTGAGCTCCGTTAAAATGCGAGAATAGGGTTAGTTGCTCGTCATTACCGCTCGTCGTGTTCCCGCTGGCTATCGATTGGAAACAACATGCAATGTGGCAGGATCTTGTTGGTAGTCTGTGTAGTCGTTGTCCCGGTCGGCGTTGTCGGTGTAGT
**************************************************...((((..(((((..((....))..(((((((.(((((......))).)))))))))..((((((((..............)))).))))(((((.((((((.((((((........))...)))))))))).)))))..)))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|
| .................................................................................................................GCGAGAGTAGGGTTAG......................................................................................................................... | 16 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ................................................................................................................................................................................................TCAGGATCTTGTTGGT.......................................... | 16 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 |
| .................................................................................................TGATCTCCGTTAAGATG........................................................................................................................................ | 17 | 2 | 12 | 0.08 | 1 | 0 | 0 | 1 |
| ...................................................................................................................GAGAATAGCCTTAGTTG...................................................................................................................... | 17 | 2 | 13 | 0.08 | 1 | 0 | 0 | 1 |
| ..................................................................................................................................TGCTCGTCTTTACAGCT....................................................................................................... | 17 | 2 | 19 | 0.05 | 1 | 1 | 0 | 0 |
| ...........................................................................................................................................TCACCGCTCGTCGTGTT.............................................................................................. | 17 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| .................................................................................AAACTCGATTTCTCCG......................................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 |
Generated: 04/23/2015 at 09:38 PM