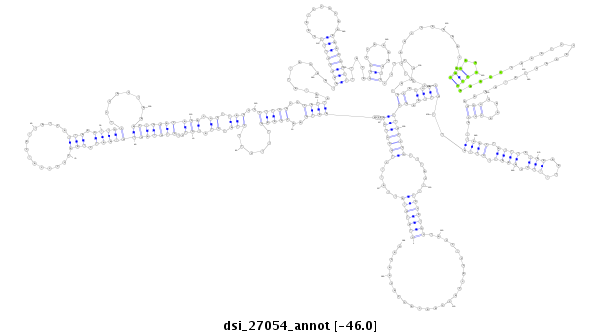
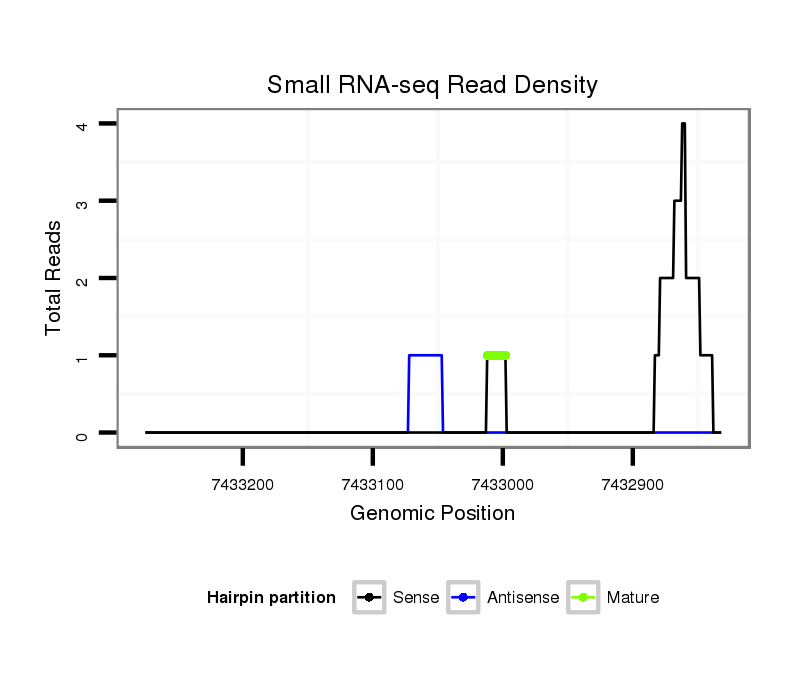
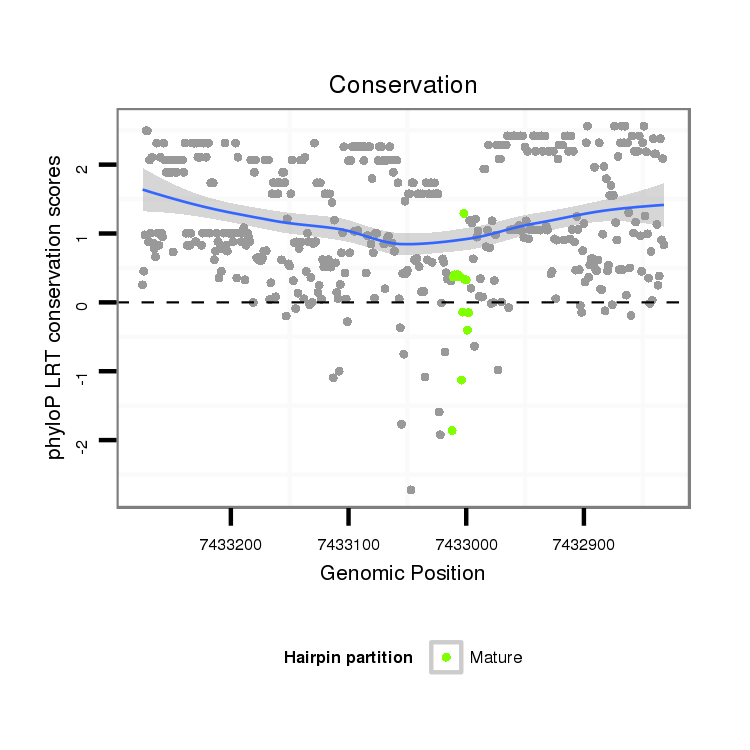
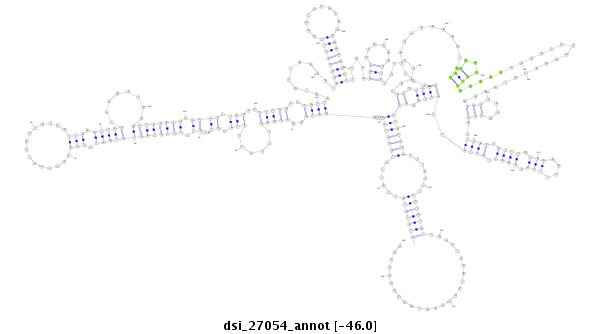
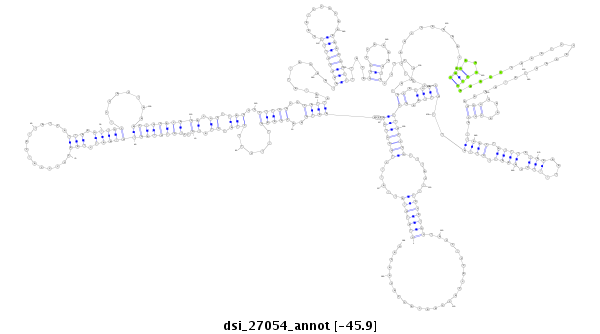
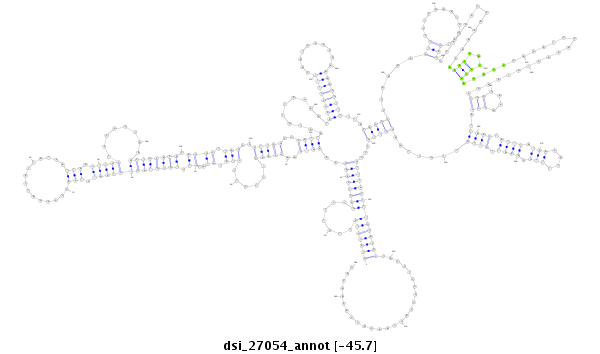
ID:dsi_27054 |
Coordinate:3r:7432882-7433225 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -46.0 | -45.9 | -45.7 |
 |
 |
 |
intergenic
| Name | Class | Family | Strand |
| TART_DV | LINE | Jockey | - |
|
ACCAACTTCATCATCCAAATCAGCACCACCAAAATCATCTCTTTAGAGACGTATGTATTATTCTGGGTGAACCAAATGAACCAAGTTTCTTCTATGTATGTCTGTGTAGTGTAATCTAACCCCCATCATTATCATTAAGTTGCTCCACTAAACTGCACACACGCTTAGCCCTTGGCCCATTAATATTAGAATAAGCCTTTCAATCCGCAAAAAGGTTTTTACAGGCCCAAAAGTCCCTCGAATGCATGACCCTCAACCAAAACACAGAACCTGAAACCCAAATCCAAAATATAAAAGGGGAAACCCATTAACTATTTATAAAATTTTGCAAATTGTTATTGTTAAGCTTGCCCGTTTACTTTGCATGCTACCTCCACCACCAACACCACAACAGCAACAGCAACAGCAGCAGCATCAGCAGCAAGCAACAATCCAGCAGCAATC
**************************************************(((((((......(((((((....((((..(((((......(((.((.(((..((((((((((((.(((...............))).)))))........)))))))))).)).)))..)))))..))))...........((((((((..........)))))))).....(((......)))........((.((((..............(((...))).......................(((....)))..((((.((((.(((....))).)))).))))..)))).))))))))).......)))))))..........................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
O001 Testis |
SRR553486 Makindu_3 day-old ovaries |
M053 female body |
SRR553485 Chicharo_3 day-old ovaries |
GSM343915 embryo |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................................................................................................ACACTGAACCTGAAACC...................................................................................................................................................................... | 17 | 1 | 2 | 2.50 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................ACAGAACCTGAAACC...................................................................................................................................................................... | 15 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................ACAGCAACAGCAGCAGCATC............................ | 20 | 0 | 6 | 1.00 | 6 | 0 | 6 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................ATCAGCAGCAAGCAACAATCCAGC....... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................AGCAACAGCAACAGCAGCAGCATC............................ | 24 | 0 | 3 | 1.00 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................AGCAGCATCAGCAGCAAGCA................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................ACAGTACCTGAAACC...................................................................................................................................................................... | 15 | 1 | 3 | 1.00 | 3 | 1 | 0 | 1 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................ACACTGAACCTGAAAC....................................................................................................................................................................... | 16 | 1 | 7 | 0.57 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................ATAGAACCTGAAACC...................................................................................................................................................................... | 15 | 1 | 12 | 0.33 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................TTTTGCTAATTGTTATAGTT..................................................................................................... | 20 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................................................................CACTGAACCTGAAACC...................................................................................................................................................................... | 16 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................AAACAGTACCTGAAACC...................................................................................................................................................................... | 17 | 2 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................CTATGTCTGTGTAGT.............................................................................................................................................................................................................................................................................................................................................. | 15 | 1 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................................................................................................................................................................................GACACTGAACCTGAAACC...................................................................................................................................................................... | 18 | 2 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................CACTGAACCTGAAAC....................................................................................................................................................................... | 15 | 1 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................CACTGAACCTGAAACCA..................................................................................................................................................................... | 17 | 2 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................ACACTGAACCTGAAACCA..................................................................................................................................................................... | 18 | 2 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................TCTTTGTGGAGTGTAATCTA...................................................................................................................................................................................................................................................................................................................................... | 20 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................GTCTGGGTAGTGTAGTC........................................................................................................................................................................................................................................................................................................................................ | 17 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................................................................................................................................................AAACAGAACCTGAAA........................................................................................................................................................................ | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................ACAGCACCTGAAACCGAA................................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TAGTCGGGGTAAACCAAAT............................................................................................................................................................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................................................................AACAGTACCTGAAACC...................................................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TGGTTGAAGTAGTAGGTTTAGTCGTGGTGGTTTTAGTAGAGAAATCTCTGCATACATAATAAGACCCACTTGGTTTACTTGGTTCAAAGAAGATACATACAGACACATCACATTAGATTGGGGGTAGTAATAGTAATTCAACGAGGTGATTTGACGTGTGTGCGAATCGGGAACCGGGTAATTATAATCTTATTCGGAAAGTTAGGCGTTTTTCCAAAAATGTCCGGGTTTTCAGGGAGCTTACGTACTGGGAGTTGGTTTTGTGTCTTGGACTTTGGGTTTAGGTTTTATATTTTCCCCTTTGGGTAATTGATAAATATTTTAAAACGTTTAACAATAACAATTCGAACGGGCAAATGAAACGTACGATGGAGGTGGTGGTTGTGGTGTTGTCGTTGTCGTTGTCGTCGTCGTAGTCGTCGTTCGTTGTTAGGTCGTCGTTAG
**************************************************(((((((......(((((((....((((..(((((......(((.((.(((..((((((((((((.(((...............))).)))))........)))))))))).)).)))..)))))..))))...........((((((((..........)))))))).....(((......)))........((.((((..............(((...))).......................(((....)))..((((.((((.(((....))).)))).))))..)))).))))))))).......)))))))..........................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
GSM343915 embryo |
SRR618934 dsim w501 ovaries |
M025 embryo |
SRR902009 testis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................TCGGGAGCCGGGTAATCAT................................................................................................................................................................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................AGGCGTTTTTCCAAAAATGTCCGGGT....................................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................................GGGGTGGTGGTTGTGG......................................................... | 16 | 1 | 20 | 0.25 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................TGGAGTTGTGGGTTGTGGTGT...................................................... | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................TCGTTCTAGTTGTCGTTCGTTGT.............. | 23 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................CGTTCTAGTTGTCGTTCGTTGT.............. | 22 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................TGGGTGCAGGTTTTATA........................................................................................................................................................ | 17 | 2 | 17 | 0.12 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................GGGTGGTGGTTGTGG......................................................... | 15 | 1 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................GTTGTCGTTGTCGTTGTCGT.................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................................................................................CAAAAATGTCTGGATTTTAA.................................................................................................................................................................................................................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................TCGTCGTAGTCGTCG...................... | 15 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................................................................................................AGCTTGCATGCTGGGAGT............................................................................................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................AAGGTTGTATCTTTTCCCC................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................GGAGCTGGCTAATTATAAT................................................................................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 3r:7432832-7433275 - | dsi_27054 | ACCAACTTCATCATCCAAATCAGCACCACCAAAATCATCTCTTTAGAGACGTATGTATTATTCTGGGTG--AACCAAATGAACCAAG----TTTCTTCTATGT--ATGTCTGTG-------TAGTGTAATCT--AACCC--------------------------------------------C------------CATCATTATCATTA----A-----------------GTTGCTCCA------CTAAACTGCACACACGC--------TTAG--------------CC--C------TTGGCCCATTAATATTAGA---ATAAG-------CCTTTCAATCCGCAAAAAGGTTTTTA-CAGGCCCA-AAAGT-CCCTC------GAAT--GCATGA-CCCTC---------AACCAAAAC-------ACAGAA---------CCT-GAAACCCA-------------------------AATCCA----------------------------A---------------------------------------AA--TATAAAA--GG--GGAAA---------CCCATTAACTATTTATAAAATTTTGCAAATTGTTATTGTTAAGCTT----GCCCGTTTACT-TTGCATGCTA----CCT-CC---ACCA------C------CAACACCACAAC-----------AGCAACAGCAACA-------------GCAGCAGCATCAGCAGCAA-----GCAACAATCCAGCAGCAATC |
| droSec2 | scaffold_12:318640-319092 - | ACCAACTTCATCATCCAAATCAGCACCACCAAAATCATCTCTTTAGAGACGTATGTATTATTCTGGGTG--AACCAAATGAACCAAG----TTTCTTCTATGT--ATGTCTGTG-------TAGCGTAATCT--AACCC--------------------------------------------C------------CATCATTATCATTA----A-----------------GTTGCTCCA------CTAAACTGCACACACGC--------TTAG--------------CC--C------TTGGCCCATTAATATTAGA---ATAAG-------CCTTTCAATCCGCAAAAAGGTTTTTA-CAGGCCCG-AAAGG-CCCACGT----ATAT--GCATGA-CCCCA---------AACCAAAACCAAGAGCCCAGAA---------CCT-GAAACCCA-------------------------AATCCA----------------------------A---------------------------------------AA--TATAAAA--GG--GGAAA---------CCCATTAACTATTTATAAAATTTAGCAAATTGTTATTGTTAAGCTT----GCCCGTTTACT-TTGCATGCTA----CCT-CC---ACCA------C------CAACACCACAAC-----------AGCAACAGCAACA-------------GCAGCAGCTTCAGCAGCAA-----GCAACAATCCAGCAGCAATC | |
| dm3 | chr3R:13837386-13837861 + | ACCAACTTCATCATCCAAATCAGCACCACCAAAATCATCTCTTTAGAGACGTATGTATTATTCTGGGTG--AACCAAATGAACCAAG----TTTCTTCTATGT--ATGTCTGTT-------TAGCGTAATCT--AACCC-------------------------C--------------------------------ATCATTATCATGATATAA-----------------GTTGCTCCA------CTAAACTGCACACACGC--------TTAG--------------CC--C------TTGGCCCATTAATATTAGA---ATAAG-------CCTTTCAATCCGCAAAAAGCTTTTCA-CAGGCCCA-AAAGT-CCCTCGTAGATATAT--GCATGA-CCCTA---------AACCAAAAC-------ACAGAA---------CCC-GAAACCCA-------------------------AATCCA----------------------------A---------------------------------------AATATATAAAA--GG--AGAACCCCCCCCCCCCCATTAACTATTTATAAAATTTTGCAAATTGTTATTGTTAAGCTT----GCCCGTTTACT-TTGCATGCTA----CCT-CC---ACCA------C------CAACACCACAAC-----------AACAACAACAACAAC-AACAGCAACAGCAGCAGCATCAACAGCAA-----GCAACAATCAAGCAACAATC | |
| droEre2 | scaffold_4770:7788280-7788721 - | ACCAACTTCATCATCCAAATCAGCACCACCAAAATCATCTCTTTAGAGACGTATGTATTATTCTGGGTG--AACCAAATGAACCAAG----TATCTTCTATGTCT------GTG-------TAGCGTAATCT--AACCC-------------------------T--------------------------------ATCATTATCATTA----A-----------------GTTGCTCCA------CTAAACTGCACACACGC--------TTAG--------------CC--C------TTAGCCCATTAATATTAGA---ATAAG-------CCTTTCAATCCGCAAAAAGGTTTTCA-CAGGCCCG-AAAGT-CCCTC------ACAT--GCATGA-CCCCA---------AACAAAAACCCCGAACCCAGAA---------CCC-GAAACCCA-------------------------AATCCA------------------------------------------------------------------------AATAAAAAGGG--GCAAG---------CCCATTAACTATTTATAAAATTTTGCAAATTGTTATTGTTAAGCTT----GCCCGTTTACT-TTGCATGCTA----CCT-CC---ACCA------C------CAACACCACAAC--------------------AAC-------AGCAACAGCAGCAGCATGAGCAGCAA-----GCAACAATCCAGCAGCAATC | |
| droYak3 | 3R:2431147-2431585 + | ACCAACTTCATCATCCAAATCAGCACCACCAAAATCATCTCTTTAGAGACGTATGTATTATTCTGGGTG--AACCAAATGAACCAAG----TTTCTTCTATGTCT------GTG-------TAGCGTAATCT--AACCC-------------------------T--------------------------------ATCATTACTATTA----A-----------------GTTGCTTCA------CTAAACTGCACACGCGC--------TTAG--------------CC--C------TTAGCCCATTAACATTAGA---ATAAG-------CCATTCAATCCGCAAAAAGGTTTTCA-CCGGCCCG-AAAGT-CTCTC------ATAT--GCATGAACCCCGAAACCCTG-AAC-------------CCAGAACCCAGAACACCC-GAAACCCA-------------------------AATCCA----------------------------G---------------------------------------GA-----AAAA--GG--GGAAA---------CCCATTAACTATTTATAAAATTTTGCAAATTGTTATTGTTAAGCTT----GCCCGTTTACT-TTGCATGCTA----CCT-CC---ACCA------C------CAACACCACAAC--------------------AACA-------------GCAGCAGCATGAGCAGCAA-----GCAACAATCCAGCAGCAATC | |
| droEug1 | scf7180000409246:185757-186216 + | ACCAACTTCATCATCCAAATCAGCACCACCAAAATCATCTCTTTAGAGACGTATGTATTATTCTGGGTG--AACCAAATGAACCAAG----TTTCTTCTAAGTCT------GTG-------TAGCGTAATCT--AACCC-------------------------GA-------------------------------ACCATTATCATTA--GTA-----------------GTTGCTCGA------CTTAACTGCACACACCC--------TTAC-A-CA-------CTCCTTT------TAAGCCCATAAATATTAGA---ATAAG-------CCATTCAATCCGCAAAAAGGTTTTCA-CAGGCCCAAAAGATCCCCTC------GTAT--GCATGA-CCCAC--------A----A---------------------A-A----A-AAAAGTCAAC----------------------------------------T-----CACTGGCCTTAAA---------------------------------------CAAAATAAAAACGG--GGAAC---------CCCATTAACTATTTATAAAATTTTGCAAATTGTTATTGTTAAGCTT----GCTCGTTTACT-TTGCATGCTA----CCT-C------CA------C------CAACACCACAAC-----------AACAACAACAACAATTAACAACAGCAGCAGCAACATCAGCAGCAA-----GCAACAATCCAGCAGTAATC | |
| droBia1 | scf7180000302075:3320746-3321195 + | ACCAACTTCATCATCCAAATCAGCACCACCAAAATCATCTCTTTAGAGACGTATGTATTATTCTGGGTG--AACCAAATGAACCAAG----TTTCTTCTAAGTCT------GTG-------TAGCGTAGTCT--AACCC-------------------------TAGA-TTGAGAGCC--CCAC------------CACCACTGTCATTA----------------------GTTGATCCACATCCGCTGAATTGCACACACACACACACACACAGTTATA--------CCC--T------TTACCCCATAAATATTAGA---ATAAG-------CCATTCAATCCGCAAAAAGGTTTGGC-CCGGCCCTAAAAAT-CCCTC------ATAT--GCATGA-CCCCA---------A---A--------------------------------AA-----------------------------------------------------------CGTAAA---------------------------------------CAAAATAAAA--GGACCCAAG---------CCCATTAAGTATTTATAAAATTTTGCAAATTGTTATTGTTAAGCTT----GCTCGTTTACT-TTGCATGCTA----CCT-CCTCCACCA------C------CAACACCACAAC--------------------AACA-------------GCAGCAACATCAGCGGCAA-----GCAACAGCCCAGCGGCAATC | |
| droTak1 | scf7180000415417:795387-795846 + | ACCAACTTCATCATCCAAATCAGCACCACCAAAATCATCTCTTTAGAGACGTATGTATTATTCTGGGTG--AACCAAATGAACCAAG----TTTCTTCTAAGTCT------GTG-------TAGCGTAGTCT--AACCC------------------------CTAGAATTGAGTGCT--CCAC------------CACCATTATCATTA----------------------GTTGATCCACAGCCACTAAACCACCCAC-----------------------------CCC--T------TTGGCCCATAAATATTAGA---ATAAG-------CCATTCAATCCGCAAAAAGGTTTCGGCCAGGCCCAAAAAAT-CCCTC------ATAT--GCATGA-CCCAC--------AGACAA-------------------------------AAACCCA-------------------------AAACCA---------TTC-----CGCTGGTCCTAAA---------------------------------------CAAAATAAAA--G---GCAAC---------CCTATTAAGTATTTATAAAATTTTGCAAATTGTTATTGTTAAGCTT----GCTCGTTTACT-TTGCATGCTA----CCT-CC---ACCA------C------CAACACCACAAC-----------AACAACAACAACA-------------GCAGCAACATCAGCAGCAA-----GCAACAATCCAGCAGCAATC | |
| droEle1 | scf7180000491008:2426617-2427031 - | ACCAACTTCATCATCCAAATCAGCACAACCAAAATCATCTCTTTAGAGACGTATGTATTATTCTGGGTG--AACCAAATGAACCAAG----TTTCTTCTAAGTCT------GTG-------TAGAGTAATCTCTAGCC----------------------------------------------------------CACCATAATCATTA----------------------GTTGCTCCA------CTAAACTGCACAC------------------------------CC--T------TTATCCCATAAATATTAGA---ATAAG-CT-AGGCCATTCAATCCGCAAAAAGGTTTTGGCCAGGCCCG-AAAAT-CCCCC------ACAA--GCATGA-CCCCG---------T---T---------------------A------------------------------------------------------------------------------A---------AGACCCGTTT-----------CCCCTAAA--AATATAA--G---CGAAC---------CCCATTAAGTATTTATAAAATTTTGCAAATTGTTATTGTTAAGCTT----GCCCGTTTACT-TTGCATGCTA----CCAACC---ACCA------C------CAACACCACAAC--------------------AACA-------------GCAGCAACATCAGCAGCAA-----GCAACAATCCAGCAGCAATC | |
| droRho1 | scf7180000758593:6596-7013 + | ACCAACTTCATCATCCAAATCAGCACCACCAAAATCATCTCTTTAGAGACGTATGTATTATCCTGGGTG--AACCAAATGAACCAAG----TTTCTTCTAAGTCT------GTG-------TAGAGTAATCTCAAACC----------------------------------------------------------CACCATTATCATTA----------------------GTTGCCCCA------CTAAACTGCACAC------------------------------CC--T------TTAGCCCATAAATATTAGA---GTAAG-------CCATTCAATCCGCAAAAAGGTTTTGCCCAGGCCCG-AAAAT-CCCTC------ATAT--GCATGA-CCCCG---------A---A---------------------A------------------------------------------------------------------------------A---------AGACCCAACTGCCCTACTCCACCCCAAAA--AATAAAA--G---CGAAC---------CCTATTAAGTATTTATAAAATTTTGCAAATTGTTATTGTTAAGCTT----GCCTGTTTACT-TTGCATGCTA----CCA------ACCA------C------CAACACCACAAC--------------------AACA-------------GCAGCAACATCAGCAGCAA-----GCAACAATCCAGCAGCAATC | |
| droFic1 | scf7180000453800:322208-322645 + | ACCAACTTCTTCATCCAAATCAGCACCACCAAAATCATCTCTTTAGAGACGTATGTATTATTCTGGGTG--AACCAAATGAACCAAG----TTTCTTCTAAGTCT------GTG-------TACAGTAGTCT--AACCC-------------------------TAGA-GTGAA--TC--CCTT------------CACCATTATCATTA----------------------GTTGCTCCA------CTATAGTGC-CGCACCC---------AAA--------------CC--T------TTAGCCCATAAATATTAGA---ATAAG-------CCATTCAATCCACAAAAAGGTTTTC------CCCG-AAAAT-CCCTC------ACATGCGCATGA-CCCCA---------A---A-------------------------------AAACCCA-------------------------A--CCA---------ATT-----CACAGGCCCCAAAAA---------AGA-----------------------AAA--AATTAAA--G---CGAAC---------CCCATTAACTATTTATAAAATTTTGCAAATTGTTATTGTTAAGCTT----GCTCGTTTACT-TTGCATGCTA----CCA------ACCA------C------CAACACCACAAC--------------------AACA-------------GCAGCAACATGAGCAGCAA-----GCAACAATCCAGCAGCAATC | |
| droKik1 | scf7180000302809:457409-457864 - | ACCAACTTCATCATCCAAATCAGCACCACCAAAATCATCTCTTTAGAGACGTATGTATTATTCTGGGTG--AACCAAATGAACCAAG----TTTCTTCTATGTCTATGTCTGTGTGCCGTGTACAGTAATCT--AACCC-------------------------TAGA-TTGAG--CC-CCCAC------CCCG---CCCATTGTCATTA----------------------GTTGCTTCA------CTA-ACTGCACAC------------------------------TC--T------TTAGCCCATAAATATTAGA---ATAAG-------CCATTCAATCCGCAAAAAGGTTT-CC-CGGGCCCA-AAAAT-CCCTC------AAAT--GCATGA-CCCGA--------------------------------------------------------------------------------CCACATTC-CTTATT-----C-CC-----------GGCCCCGTG--------TT-----------CCCCGAAA--AATAAAA--A---CGAAC---------CCCATTAACTATTTATAAAATTTTGCAAATTGTTATTGTTAAGCTT----GCTCGTTTACT-TTGCATGCTA----CC--CC---ACCACCACCAC------CAACAACACCAC--------------------AGCA-------------GCAGCAACATGAGCAGCAA-----GCAACTACCCAGCAGCAATC | |
| droAna3 | scaffold_13340:11879011-11879484 - | ACCAACTTCATCATCCAAATCAGCACCACCAAAATCATCTCTTTAGAGACGTATGTATTATTCTGGGTG--AACCAAATGAACCAAG----TTACTTCTAAGTCT------GTGTACCG--TACAGTAATCT--AACCC-------------------------TATA-T-AAA--CCCCCCACCGTAACACCGGTCACCATTGTCATTA----------------------GTTGCTCCA------CTA-ACTGCACACACT----------------------------C--T------TTAGCCCATAAATATTAGA---ATAAG-------CCATTCAATCCGCAAAAAGGTTTTCT-TCGACCCG-AAAAT-CCCTC------ATAT--GCACGA-CCCGA---------A-------------------------AAA-----------------------------------------T----GCTCCCATGCC-----G-------------A---------AGAGCC-------------------AAAAAATATAAAA--A---CGAAC---------CCCATTAACTATTTATACAATTTTGCAAATTGTTATTGTTAAGCCT----GCTCGTTTACT-TTGCATGCAAACCACCA-TC---ACCA------CCAACAA-----CAACTACACCACTTCCACAAC------AAC-------ATCAGCAGCAGCAACATGAGCAGCAA-----GCAACAACCAAGCAGCAATC | |
| droBip1 | scf7180000396691:768769-769250 - | ACCAACTTCATCATCCAAATCAGCACCACCAAAATCATCTCTTTAGAGACGTATGTATTATTCTGGGTG--AACCAAATGAACCAAG----TTACTTCTAAGTCT------GTGTACCG--TACAGTAATCT--AACCC-------------------------TATA-A-GAA--CCCCCTAACGTCACACCGCTCACCATTATCATTA----------------------GTTGCTCCA------CTA-ACTGCACACACT----------------------------C--T------TTAGCCCATAAATATTAGA---ATAAG-------CCATTCAATCCGCAAAAAGGTTTTCC-TCGAACCC-AAAAT-CCCTC------ATAT--GCACGA-CCCGA---------A-------------------------GAA-----------------------------------------C----ACTCCCATATG-----G-------------AG-GAGGGAGAGACCC-------------------AAAAAATATAAAA--A---CGAAC---------CCCATTAACTATTTATAAAATTTTGCAAATTGTTATTGTTAAGCCC----GCTCGTTTACT-TTGCATGCAAACCACCA-TC---ACCA------CCAACAA-----CAACTACACCACTTCCACAAC------AAC-------AACAACAGCAGCAACATGAGCAGCAA-----GCAACAACCAAGCAGCAATC | |
| dp5 | 2:28890170-28890444 + | CACAA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACAC--------TTAGTTC-A--------CCC--T------TTGACCCATAAATATTAGATGCAAAAACCT----CCATTCAATCTACCGA------------------------------------------------------C--------A----A---------------------A-A----AAAAGGGTCCACAAACCAATCCTCTAAAGCATGAGA--C------C-CTTTTTACTAAC-CT-----------GGC--------------------------------------ATAAAG--C---CGTA-----CAGACCCCATTAAATATTTATAAAATTTTGCAAATTGTTATTGTTAAGCTTTGGTGCTCGTTTACTTTTGCATGCTA----CTC-GT---ACCA------CCTACAA-----CCACTCG-----------AACCACAGCAACA-------------ACAACAGCAGCAGCAGCAT-----TCAAC--------------- | |
| droPer2 | scaffold_6:4217484-4217757 + | CACAA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACAC--------TTTGTTC-A--------CCC--T------TTGACCCATAAATATTAGATGCAAAAACCT----CCATTCAATCTACCGA------------------------------------------------------C--------A----A---------------------A-A----A-AAGGGTCCACAAACCAATCCTCTAAAGCATGAGA--C------C-CTTTTTACTAAC-CT-----------GGC--------------------------------------ATAAAG--C---CGTA-----CAGCCCCCATTAAATATTTATAAAATTTTGCAAATTGTTATTGTTAAGCTTTGGTGCTCGTTTACTTTTGCATGCTA----CTC-GT---ACCA------CCTACAA-----CCACTCG-----------AACCACAGCAACA-------------ACAACAGCAGCAGCAGCAT-----TCAAC--------------- | |
| droWil2 | scf2_1100000004902:4853371-4853624 - | ACCAACTTC---ATCCAAATCAGCACCACCAAAATCATCTCTTTAGAGACGTATGTAT----ATGAGCAACAACAAAATGAATCAAGAAATCTTCCTCTATGTAT------ATG-------TAGA-----------CCCTAACTATCTATCTATCTATCTAACACAGA--------------------------------AATATGCTTA--TTCTTTCTACCACTTTAATTGCTGACCCA------C------------------------TTAT-A-TTTTGCCCATCCGTTTTTAAAGTAATCGCATAAATATTAGA---TTAAG-CCGAC--GCAACAATCCACAAAAGGGTT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droVir3 | scaffold_12723:5779719-5779849 - | TA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A-CAATTTTAAACTGTTGTTGTTTGCTGCTGTTAGGCGC----GTC------AT-TTGTAAGCCG----CTT-AC---GCCA------C------AGTAATAACAGC-----------AACAACAGCAAAA-------------ACAACAACAACAGCAGCAACAACAACAACAATTAAGCAACA--- | |
| droMoj3 | scaffold_6540:2703527-2703671 - | AA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AA---GG--CGAAG---------CCAATTAACTATTTATGAAAT--TGCAAATTGTTATTGTTTAGCTCG---GCTCGTTTACT-TTGCATGGCA----AC-------AGCA------G------CAACACAGCAAC-----------AGCAACAGCAACA----------ACAGCAGCAGCAACAGCAGCA------GCAA-----CAGCAAAAATT | |
| droGri2 | scaffold_15245:2175401-2175572 + | TCAAAT-ACAGAACACGCA-----------------ACAAACTTAAAGACTAAAGTAC----AAATTAAACTAAAGGGCAGAACTTAAAAGTTGATGT----------------------------------------------------------------------------------------------------------------------------CACGCAAATTGACAGCACA------T------------------------TTCC-G-TGGCAATCTCCA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A----CC--CC---AGCA------GCAGCAG-----CAGCAGC-----------AGCAGCAGCA----------------GCAGCAGCA---------------GCAGCAG--CAGCAGCAG-C |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/18/2015 at 11:45 AM