ID:dsi_26861 |
Coordinate:3l:15987196-15987267 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
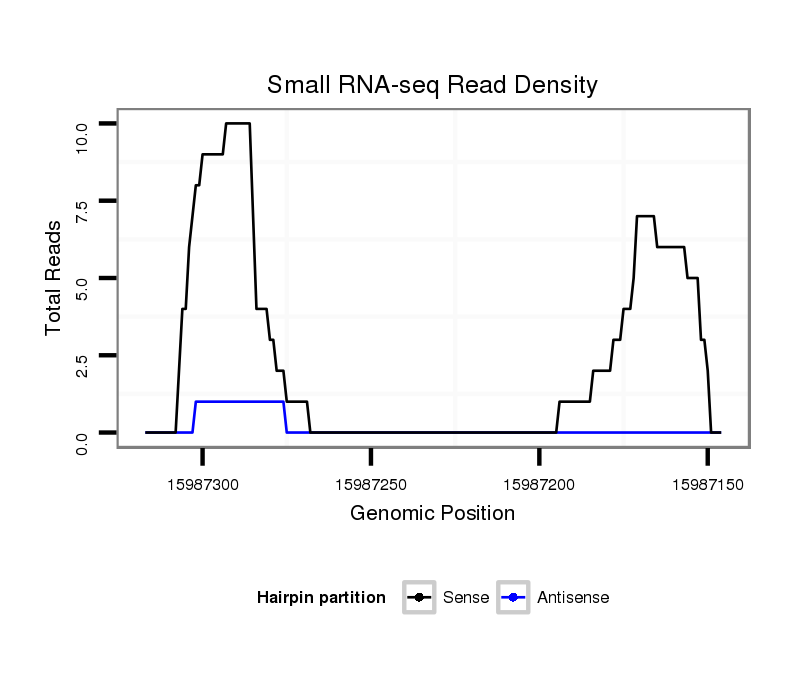
| Legend: | mature | star | mismatch in read |
 |
exon [3l_15986838_15987195_-]; CDS [3l_15986838_15987195_-]; CDS [3l_15987268_15987803_-]; exon [3l_15987268_15987803_-]; intron [3l_15987196_15987267_-]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| ##################################################------------------------------------------------------------------------################################################## TCTGTCCTGCCATCTACGACATCAATCTGGCCTTCAAGAAGAATGCCGAGGTGAGAACTTAAGATCTCATATAAATATATATAAATTTTAATTTACATTTTGCAACTTGAATTGATTTACAGCCTAAGCCCACCATGCTGTCCCAGTTGAACGGCGAGCCCGTGGAACCGTA **************************************************.......((...(((((((............(((((....)))))............)))...))))...))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
M053 female body |
SRR618934 dsim w501 ovaries |
GSM343915 embryo |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
M024 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................TGCGGTACCAGTTGAACGGCGC............... | 22 | 3 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ..........CATCTACGACATCAATCTGGCC............................................................................................................................................ | 22 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........ATCTACGACATCAATCTGGCCT........................................................................................................................................... | 22 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TTGAACGGCGAGCCCGTGG....... | 19 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................TGGCCTTCAAGAAGAATGCCGAGC......................................................................................................................... | 24 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................CCTGTTGAACGGCGAAACCGTGG....... | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................CGAGCCTAAGCCCACCATGCT................................. | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............TACGACATCAATCTGGCCTTCAAGA..................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............CTACGACATCAATCTGGCCTTCAA....................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................ATCTGGCCTTCAAGAAGAATGCCGA........................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................CATGCTGTCCCAGTTGAACGGCGAGCCC........... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................GTTGAACGGCGAGCCCGTGGAAC.... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................GTCCCAGTTGAACGGCGAGCCCGTGGAA..... | 28 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................CTGCCGTTCAAGAAGCATGCCG............................................................................................................................ | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............CTACGACATCAATCTGGCC............................................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................CCAGTTGAACGGCGAGCCCGTGGAAC.... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................GACATCAATCTGGCCTTCAAGAAGA.................................................................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............ACGACATCAATCTGGCCT........................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................CTAAGCCCACCATGCTGTCCCAGTTGAAC.................... | 29 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................AGAATGCGTAGGTGAGGAC.................................................................................................................. | 19 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
AGACAGGACGGTAGATGCTGTAGTTAGACCGGAAGTTCTTCTTACGGCTCCACTCTTGAATTCTAGAGTATATTTATATATATTTAAAATTAAATGTAAAACGTTGAACTTAACTAAATGTCGGATTCGGGTGGTACGACAGGGTCAACTTGCCGCTCGGGCACCTTGGCAT
**************************************************.......((...(((((((............(((((....)))))............)))...))))...))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
M023 head |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................CTGGTACGACAGGGTCA......................... | 17 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............TGCTGTAGTTAGACCGGAAGTTCTTCT.................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................ACGGCTCCTCTCTTGAA................................................................................................................ | 17 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................CGGCTCCTCTCTTGAA................................................................................................................ | 16 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| AGACAGTACGGTAGA............................................................................................................................................................. | 15 | 1 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................CTTCTTACGGCTACA........................................................................................................................ | 15 | 1 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................AGATTATATTTATATAAATTTAAT.................................................................................... | 24 | 3 | 19 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................GCCGGAATTGCTTCTTAC............................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................................................................AGACTATATTTATTTATATTT....................................................................................... | 21 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 12:17 AM