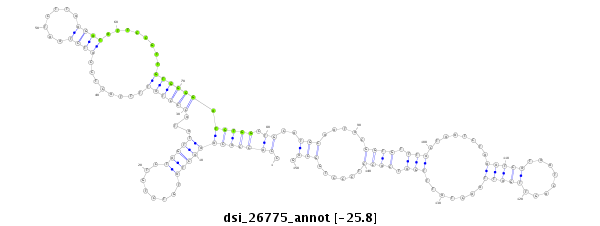
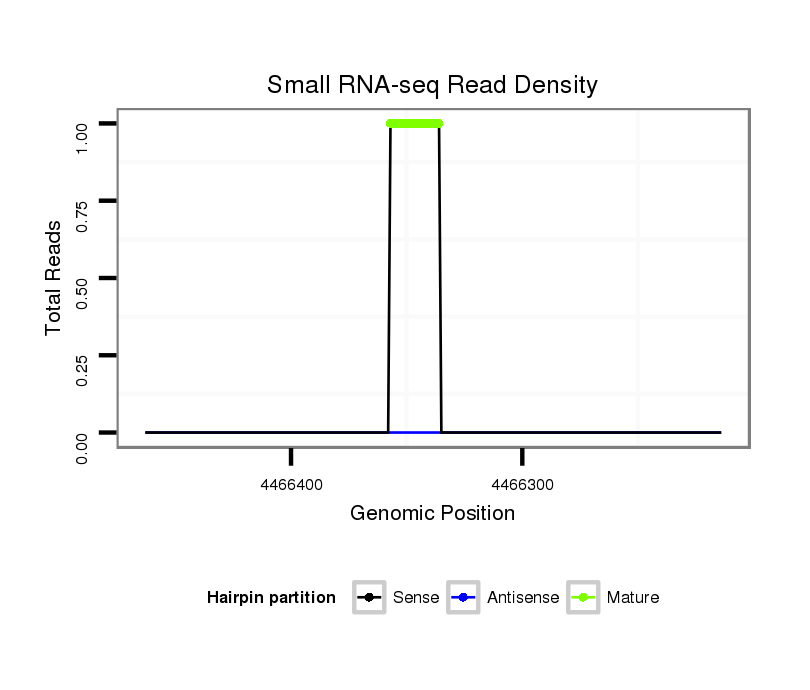
ID:dsi_26775 |
Coordinate:3r:4466264-4466413 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -25.7 | -25.7 | -25.6 |
 |
 |
 |
exon [3r_4466115_4466263_-]; CDS [3r_4466115_4466263_-]; intron [3r_4466264_4469295_-]
No Repeatable elements found
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## GCTTTAATCTCTCTTTTTTATCTATTTCCCGTCACCACTTCGTTTGTATGCCACCACAAACTATGTCTGTCTAGTTATACCCTATTTTACTCCATCTAATCTTAAGATATTCCGTTGTGGGGATGTGGCTGCATGCAATACGGTGTTTACTAATCTAAATCATAATCAGTTGATTAACTATTTGATCACCTCCCTGGCAGACGATGAATTGGCAGCCAAAATCAAGGAGGTCGAGAAAAAACCCGTTCAG **************************************************.((((((((((((........)))))...((((((........((((.......))))........)))))).))))).))..(((.....((((.(((........(((((........))))).......))).))))......))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902008 ovaries |
M053 female body |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR902009 testis |
GSM343915 embryo |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................................TCAAGGAGCTCGAGAAAAA.......... | 19 | 1 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................ATATTCCGTTGTGGGGATGTGG.......................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................GTGGCTGCATGCACTA.............................................................................................................. | 16 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................TCACCTCCCTGGCAGA................................................. | 16 | 0 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................TCAAGGAGCTGGAGAAAAAA......... | 20 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................GGAGGTCGCGAATCAACCC...... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................GAACTATGGCTGTCTAGTA.............................................................................................................................................................................. | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................CTATGTCTGTGTAGT............................................................................................................................................................................... | 15 | 1 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................ATGAGGTCGAGAAACCACC....... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................CATTCTATACGGTGTT....................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CGAAATTAGAGAGAAAAAATAGATAAAGGGCAGTGGTGAAGCAAACATACGGTGGTGTTTGATACAGACAGATCAATATGGGATAAAATGAGGTAGATTAGAATTCTATAAGGCAACACCCCTACACCGACGTACGTTATGCCACAAATGATTAGATTTAGTATTAGTCAACTAATTGATAAACTAGTGGAGGGACCGTCTGCTACTTAACCGTCGGTTTTAGTTCCTCCAGCTCTTTTTTGGGCAAGTC
**************************************************.((((((((((((........)))))...((((((........((((.......))))........)))))).))))).))..(((.....((((.(((........(((((........))))).......))).))))......))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................AAAATGAGGAAGGTTAGAATTGT............................................................................................................................................... | 23 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................AAATGAGGAAGGTTAGAATTGT............................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................AGGGACCGTCTGCTACTTTTC....................................... | 21 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................AGATTTAATATTAGTCAGC.............................................................................. | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................AGAATTCTCTAAGACAACA.................................................................................................................................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................AGAATTCTCTAAGACAAC..................................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................CTAGTCGAGGGACCGCCT................................................. | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................GAATTCTCTAAGACAAC..................................................................................................................................... | 17 | 2 | 19 | 0.11 | 2 | 1 | 0 | 0 | 0 | 1 |
| ..........................AGGGCACTGGTGCAGCAAC............................................................................................................................................................................................................. | 19 | 3 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........GGAGACAAAATAGATAAA............................................................................................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
Generated: 04/24/2015 at 08:36 AM