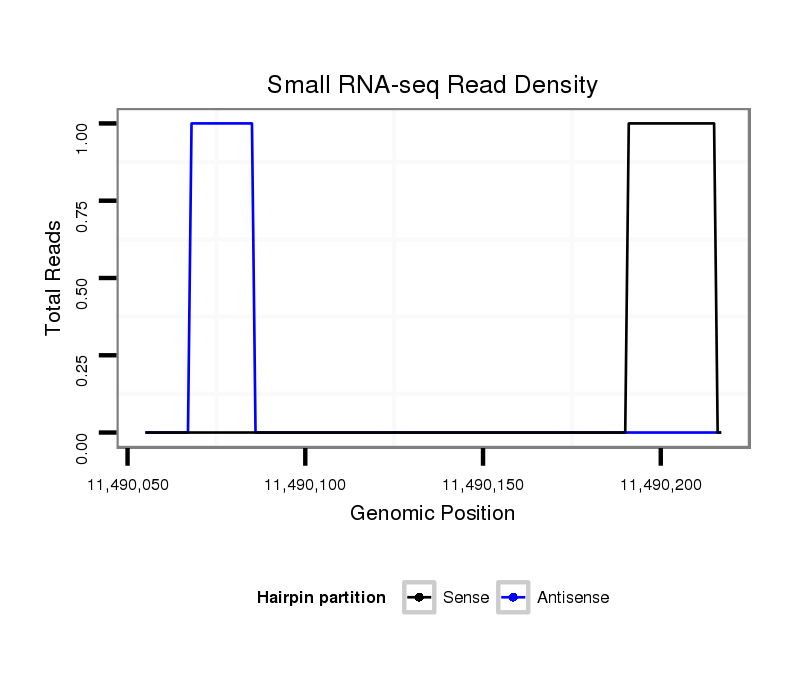
ID:dsi_26443 |
Coordinate:3l:11490105-11490167 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
exon [3l_11489898_11490104_+]; CDS [3l_11489898_11490104_+]; CDS [3l_11490168_11490495_+]; exon [3l_11490168_11490513_+]; intron [3l_11490105_11490167_+]
No Repeatable elements found
| ##################################################---------------------------------------------------------------################################################## AAGGAGAAGTACACCGCCAAAGTAAAGCTTCAACCGAAAGGGCAGTCAAGGTGGGATATAAATTTTATTCTTAAACCGCTTTTAAAGATTTGACAAATTATTTACAATTTTAGGGAGGCATTTACACTGAGCCTACTCTCCAAATTCCGAACGAAACTGGATA **************************************************((((..((..(((...)))..))..))))..................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M053 female body |
SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|
| ......AAGTACACCGCCGAAGT............................................................................................................................................ | 17 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................TCTCCAAATTCCGAACGAAACTGGA.. | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| AAGGAGAAGTACACCG................................................................................................................................................... | 16 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| ........GTACACCGCCGAAGT............................................................................................................................................ | 15 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................CTTCAACCGCAAGGG......................................................................................................................... | 15 | 1 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................TCTGACAAATTATTTAAAA........................................................ | 19 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 |
| ...................................................................................................................................................CGAGCGAAACTGGAT. | 15 | 1 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....................GTAGAGCTTCGACCGAA............................................................................................................................. | 17 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 |
| .....GAAGCACACCGCCAAT.............................................................................................................................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
|
TTCCTCTTCATGTGGCGGTTTCATTTCGAAGTTGGCTTTCCCGTCAGTTCCACCCTATATTTAAAATAAGAATTTGGCGAAAATTTCTAAACTGTTTAATAAATGTTAAAATCCCTCCGTAAATGTGACTCGGATGAGAGGTTTAAGGCTTGCTTTGACCTAT
**************************************************((((..((..(((...)))..))..))))..................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
O001 Testis |
GSM343915 embryo |
SRR902009 testis |
SRR553487 NRT_0-2 hours eggs |
M024 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................TGTGACACGGATGCGAGGTT.................... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................ATCGCTCCGTCCATGTGACTC................................ | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............GGCGGTTTCATTTCGAAG.................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................AATGGGAGGTTTAAGGCT............. | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................TCCCGTCAGCTCCAC.............................................................................................................. | 15 | 1 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................CGCGGCTGAGAGGTGTAAG................ | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 10:21 AM