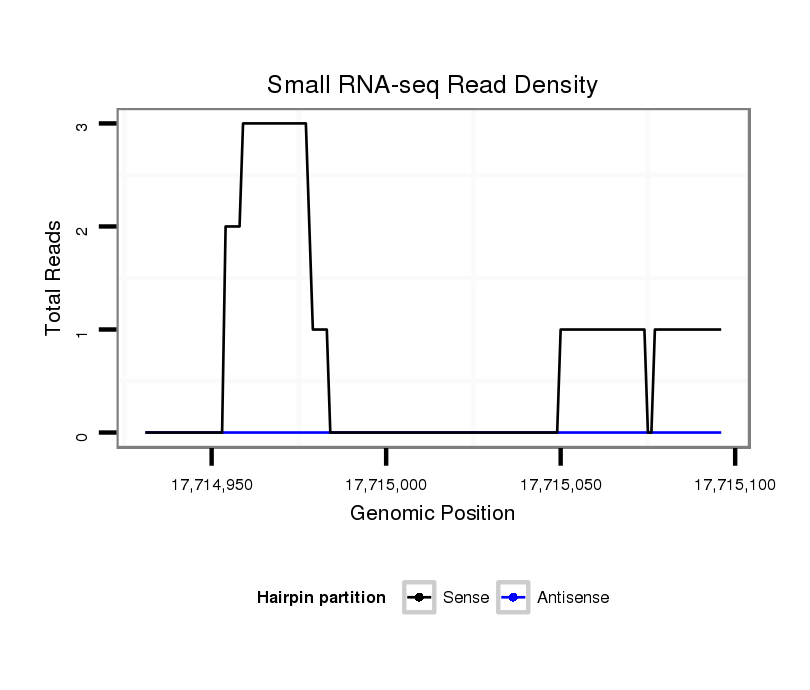
ID:dsi_26313 |
Coordinate:3r:17714981-17715046 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
CDS [3r_17715047_17715217_+]; exon [3r_17714700_17714980_+]; five_prime_UTR [3r_17715047_17715222_+]; exon [3r_17715047_17715820_+]; CDS [3r_17714700_17714980_+]; exon [3r_17715047_17715820_+]; five_prime_UTR [3r_17714700_17714980_+]; exon [3r_17714700_17714980_+]; intron [3r_17714981_17715046_+]; intron [3r_17714981_17715046_+]
No Repeatable elements found
| ##################################################------------------------------------------------------------------################################################## TGCTGGACGGGGTCAACGCACTCTTGGCAAAAGCGAAGGTGGAACGAACTGTTAGTGGGTCAGCCTAATCTCCGCCTGTTTATCATTGATTAATTGGTCTTCTGTTCAAATTGCAGGCCAGCACCCGGTTGCAGGAGGCGCGACTGGCACGTGAGCAACGCCGGGC **************************************************.........................((((.................................))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902009 testis |
SRR618934 dsim w501 ovaries |
M024 male body |
M053 female body |
SRR902008 ovaries |
SRR553485 Chicharo_3 day-old ovaries |
M025 embryo |
SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................GCACGTGAGCAACGCCGGGC | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................TTGGCAAAAGCGAAGGTGGAACGAA...................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................TGCCAGCACCCGGTTGCAGGAGGCGCGAC...................... | 29 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................TGGCAAAAGCGAAGGTGGAACGAACAA................................................................................................................... | 27 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................TTGGCAAAAGCGAAGGAGGAACGAA...................................................................................................................... | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................AGCACCCGGTTGCAGGAGGCGCGAC...................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................TTGGCAAAAGCGAAGGTGGAACGA....................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................AAAAGCGAAGGTGGAACGAACTGTT................................................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .CCTGGACGGGGTCAACG.................................................................................................................................................... | 17 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................GTTGAACGAACTTTTAGT.............................................................................................................. | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................CTGTTAGTGGGTGATCCT.................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................ATAGCGCCCGGTTGCAGGA.............................. | 19 | 3 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................CCCGTTGGCAGGAGGCGTGA....................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................................AGGTCGAACGAACTCTTCG............................................................................................................... | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
ACGACCTGCCCCAGTTGCGTGAGAACCGTTTTCGCTTCCACCTTGCTTGACAATCACCCAGTCGGATTAGAGGCGGACAAATAGTAACTAATTAACCAGAAGACAAGTTTAACGTCCGGTCGTGGGCCAACGTCCTCCGCGCTGACCGTGCACTCGTTGCGGCCCG
**************************************************.........................((((.................................))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
M024 male body |
|---|---|---|---|---|---|---|---|
| ......TGGCCCAGTTGCGTG................................................................................................................................................. | 15 | 1 | 2 | 0.50 | 1 | 1 | 0 |
| ................................................................................................CAGAAGGCAATTTTGACGT................................................... | 19 | 3 | 17 | 0.18 | 3 | 0 | 3 |
| ......................................................................................................................................CTCCGCGCTGGCCCTGC............... | 17 | 2 | 11 | 0.09 | 1 | 1 | 0 |
Generated: 04/24/2015 at 03:04 AM