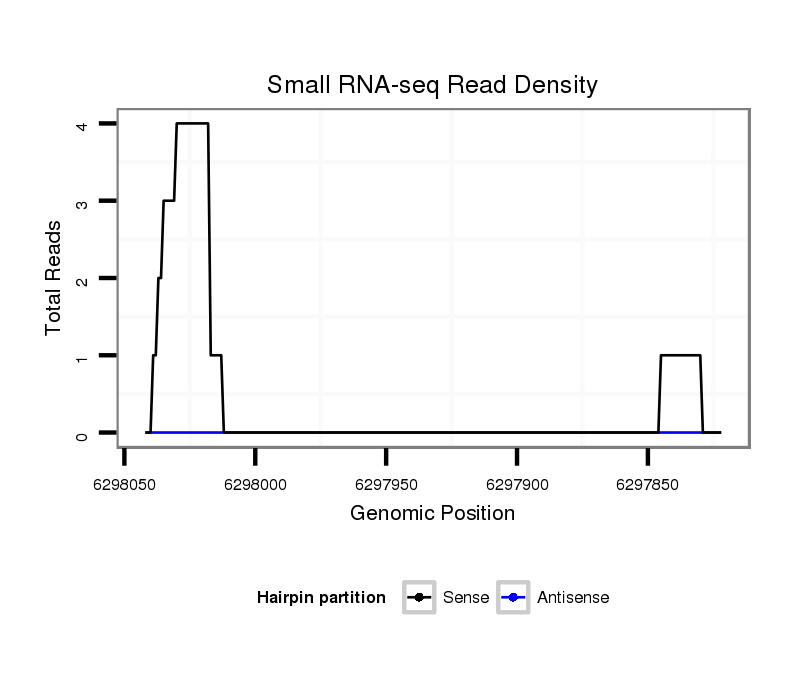
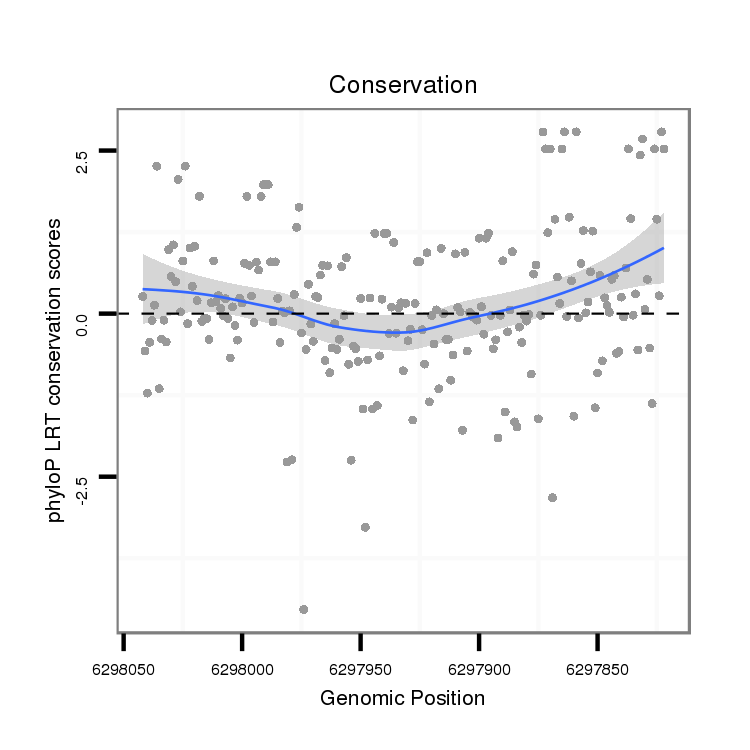
| TATCTATAATTCAGGCGCAAAAAGCAGCCCTCTCTGTCAGCAGTCCGCTGGTAAATATAT-----AGTATACATATAGATCTG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------A----------------------------------------------------------------TAT-----CACGGT------------------------------------------------GATATACGT-------------------------------TTTT-GAA--TC-TCTTGCAGCGTTGGCAGACCACAGCAGCTGTAGC------------------------------AGAGCCCAGAAATGATGAAGAGTG | Size | Hit Count | Total Norm | Total | M022
Male-body | M040
Female-body | M059
Embryo | M062
Head | V043
Embryo | V051
Head | V112
Male-body | SRR902010
Ovary | SRR902011
Testis | SRR902012
CNS imaginal disc |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................................................GACCACAGCAGCTGTAGC------------------------------AGA..................... | 21 | 3 | 1.00 | 3 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...................AAAAGCAGCCCTCTCTGTCA.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................GTTGGCAGACCACAGCAGCT........................................................... | 20 | 3 | 0.67 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................................................GACCACAGCAGCTGTAGC------------------------------AGAGC................... | 23 | 3 | 0.67 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................GTTGGCAGACCACAGCAGCTGTA........................................................ | 23 | 3 | 0.67 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACCACAGCAGCTGTAGC------------------------------AGA..................... | 20 | 3 | 0.67 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GCCCAGAAATGATGAAGAGTG | 21 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................................................CGTTGGCAGACCACAGCAGCTGT......................................................... | 23 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGAGCCCAGAAATGATGAAGA... | 21 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGACCACAGCAGCTGTAGC------------------------------AGA..................... | 22 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAGCAGCTGTAGC------------------------------AGAGCCCA................ | 21 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CCCAGAAATGATGAAGAGC. | 19 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGCCCAGAAATGATGAAGAGT. | 21 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................................................CGTTGGCAGACCACAGCAGCTGTAGC------------------------------AG...................... | 28 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GC------------------------------AGAGCCCAGAAATGATGAAG.... | 22 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGACCACAGCAGCTGTAGC------------------------------AG...................... | 21 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGC------------------------------AGAGCCCAGAAATGATGAAG.... | 23 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................................................................CCACAGCAGCTGTAGC------------------------------AGA..................... | 19 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGC------------------------------AGAGCCCAGAAATGATGA...... | 21 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGACCACAGCAGCTGTAGC------------------------------AGAGCCCAG............... | 28 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GTAGC------------------------------AGAGCCCAGAAATGATGAAGA... | 26 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................GTTGGCAGACCACAGCAGCTGTAGC------------------------------AGA..................... | 28 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GC------------------------------AGAGCCCAGAAATGATGAAGA... | 23 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGCTGTAGC------------------------------AGAGCCCAGA.............. | 19 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGC------------------------------AGAGCCCAGAAATGATGAAGA... | 24 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAGCAGCTGTAGC------------------------------AGAGCCC................. | 20 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGCCCAGAAATGATGAAGA... | 19 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................GTTGGCAGACCACAGCAGCTGT......................................................... | 22 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GTAGC------------------------------AGAGCCCAGAAATGATGAAGAGT. | 28 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................................................GGCAGACCACAGCAGCTGTAGC------------------------------AGA..................... | 25 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |